4OY2
 
 | | Crystal structure of TAF1-TAF7, a TFIID subcomplex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7, ... | | Authors: | Bhattacharya, S, Lou, X, Rajashankar, K, Jacobson, R.H, Webb, P. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insight into TAF1-TAF7, a subcomplex of transcription factor II D.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7N87
 
 | |
8IWV
 
 | |
8IX8
 
 | |
8IXX
 
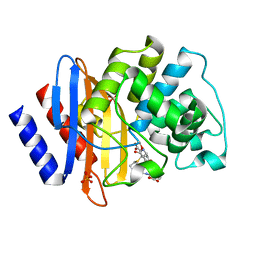 | | Crystal structure of Class A beta-lactamase BlaA WT - complex with Ertapenem | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Bhattacharya, S, Hazra, S. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Class A beta-lactamase BlaA WT - complex with Ertapenem
To be published
|
|
8IY4
 
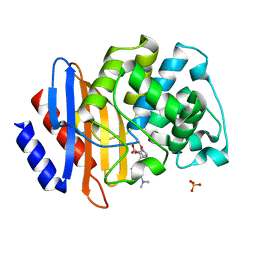 | | Crystal structure of Class A beta-lactamase BlaA WT - complex with Meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Bhattacharya, S, Hazra, S. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Class A beta-lactamase BlaA WT - complex with Meropenem
To be published
|
|
8CMV
 
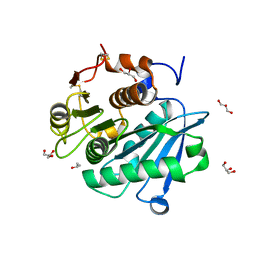 | | Engineered PETase enzyme from LCC - C09 mutant | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Leaf-branch compost cutinase | | Authors: | Bhattacharya, S, Estiri, H, Castagna, R, Parisini, E. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Development of a highly active engineered PETase enzyme for polyester degradation
Biorxiv, 2024
|
|
2NXP
 
 | |
1YSM
 
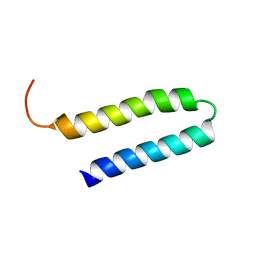 | | NMR Structure of N-terminal domain (Residues 1-77) of Siah-Interacting Protein. | | Descriptor: | Calcyclin-binding protein | | Authors: | Bhattacharya, S, Lee, Y.T, Michowski, W, Jastrzebska, B, Filipek, A, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2005-02-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Modular Structure of SIP Facilitates Its Role in Stabilizing Multiprotein Assemblies.
Biochemistry, 44, 2005
|
|
6LXP
 
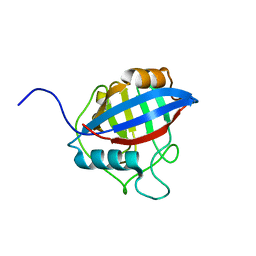 | | TvCyP2 in apo form 2 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Aryal, S, Chen, C, Hsu, C.H. | | Deposit date: | 2020-02-11 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | N-Terminal Segment of TvCyP2 Cyclophilin fromTrichomonas vaginalisIs Involved in Self-Association, Membrane Interaction, and Subcellular Localization.
Biomolecules, 10, 2020
|
|
6LXO
 
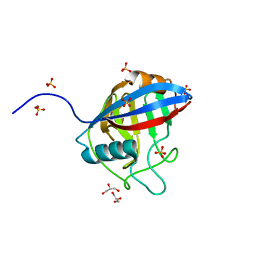 | | TvCyP2 in apo form 1 | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase, SULFATE ION | | Authors: | Aryal, S, Chen, C, Hsu, C.H. | | Deposit date: | 2020-02-11 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | N-Terminal Segment of TvCyP2 Cyclophilin fromTrichomonas vaginalisIs Involved in Self-Association, Membrane Interaction, and Subcellular Localization.
Biomolecules, 10, 2020
|
|
6LXR
 
 | | TvCyP2 in apo form 4 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Aryal, S, Chen, C, Hsu, C.H. | | Deposit date: | 2020-02-11 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | N-Terminal Segment of TvCyP2 Cyclophilin fromTrichomonas vaginalisIs Involved in Self-Association, Membrane Interaction, and Subcellular Localization.
Biomolecules, 10, 2020
|
|
6LXQ
 
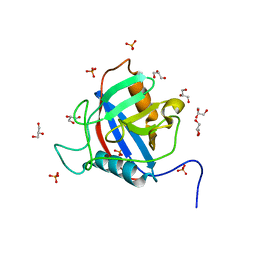 | | TvCyP2 in apo form 3 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase | | Authors: | Aryal, S, Chen, C, Hsu, C.H. | | Deposit date: | 2020-02-11 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N-Terminal Segment of TvCyP2 Cyclophilin fromTrichomonas vaginalisIs Involved in Self-Association, Membrane Interaction, and Subcellular Localization.
Biomolecules, 10, 2020
|
|
4O8G
 
 | | Structure of Infrared Fluorescent Protein 1.4 | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, 3-[2-[(Z)-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E,4R)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | Authors: | Bhattacharya, S, Forest, K.T. | | Deposit date: | 2013-12-27 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Origins of fluorescence in evolved bacteriophytochromes.
J.Biol.Chem., 289, 2014
|
|
1PSB
 
 | | Solution structure of calcium loaded S100B complexed to a peptide from N-Terminal regulatory domain of NDR kinase. | | Descriptor: | Ndr Ser/Thr kinase-like protein, S-100 protein, beta chain | | Authors: | Bhattacharya, S, Large, E, Heizmann, C.W, Hemmings, B, Chazin, W.J. | | Deposit date: | 2003-06-21 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ca(2+)/S100B/NDR Kinase Peptide Complex: Insights into S100 Target Specificity and Activation of the Kinase.
Biochemistry, 42, 2003
|
|
7S5J
 
 | |
2WAL
 
 | | Crystal Structure of human GADD45gamma | | Descriptor: | GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE PROTEIN GADD45 GAMMA, MALONIC ACID | | Authors: | Bhattacharya, S, Mueller, J.J, Roske, Y, Turnbull, A.P, Quedenau, C, Goetz, F, Buessow, K, Heinemann, U. | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Human Gadd45Gamma
To be Published
|
|
2L6E
 
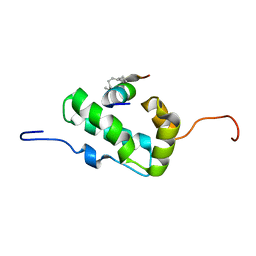 | | NMR Structure of the monomeric mutant C-terminal domain of HIV-1 Capsid in complex with stapled peptide Inhibitor | | Descriptor: | Capsid protein p24, NYAD-13 stapled peptide inhibitor | | Authors: | Bhattacharya, S, Zhang, H, Debnath, A.K, Cowburn, D. | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domain of HIV-1 capsid.
J.Biol.Chem., 283, 2008
|
|
2KRG
 
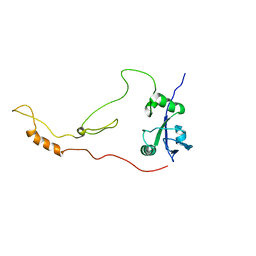 | | Solution Structure of human sodium/ hydrogen exchange regulatory factor 1(150-358) | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Dai, Z, Li, J, Baxter, S, Callaway, D.J.E, Cowburn, D, Bu, Z. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation.
J.Biol.Chem., 285, 2010
|
|
2KJD
 
 | |
2M0U
 
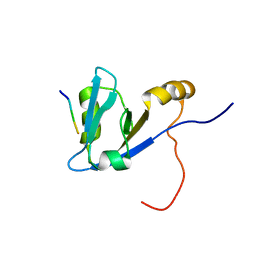 | | Complex structure of C-terminal CFTR peptide and extended PDZ1 domain from NHERF1 | | Descriptor: | C-terminal CFTR peptide, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Ju, J.H, Cowburn, D, Bu, Z. | | Deposit date: | 2012-11-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ligand-Induced Dynamic Changes in Extended PDZ Domains from NHERF1.
J.Mol.Biol., 425, 2013
|
|
2M0V
 
 | | Complex structure of C-terminal CFTR peptide and extended PDZ2 domain from NHERF1 | | Descriptor: | C-terminal CFTR peptide, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Ju, J.H, Cowburn, D, Bu, Z. | | Deposit date: | 2012-11-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ligand-Induced Dynamic Changes in Extended PDZ Domains from NHERF1.
J.Mol.Biol., 425, 2013
|
|
2M0T
 
 | |
4Z1W
 
 | | CRYSTAL STRUCTURE OF MONOMERIC BACTERIOPHYTOCHROME mutant D207L Y263F From Synchrotron | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, 3-[2-[(Z)-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E,4R)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | Authors: | Bhattacharya, S, Satyshur, K.A, Wangkanont, K, Lehtivuori, H, Forest, K.T. | | Deposit date: | 2015-03-27 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Removal of Chromophore-Proximal Polar Atoms Decreases Water Content and Increases Fluorescence in a Near Infrared Phytofluor.
Front Mol Biosci, 2, 2015
|
|
4ZRR
 
 | | Crystal Structure of Monomeric Bacteriophytochrome mutant D207L Y263F at 1.5 A resolution Using a home source. | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, 3-[2-[(Z)-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E,4R)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Bhattacharya, S, Satyshur, K.A, Lehtivuori, H, Forest, K.T. | | Deposit date: | 2015-05-12 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Removal of Chromophore-Proximal Polar Atoms Decreases Water Content and Increases Fluorescence in a Near Infrared Phytofluor.
Front Mol Biosci, 2, 2015
|
|
