8XAW
 
 | |
8XAX
 
 | |
8XAY
 
 | |
8XAV
 
 | | Cryo-EM structure of an anti-phage defense complex | | Descriptor: | ATP-binding protein, DUF4297 | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 2024
|
|
8XAU
 
 | | Cryo-EM structure of HerA | | Descriptor: | ATP-binding protein | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 2024
|
|
6TXZ
 
 | | FAB PART OF M6903 IN COMPLEX WITH HUMAN TIM3 | | Descriptor: | Fab H, Fab L, Hepatitis A virus cellular receptor 2 | | Authors: | Musil, D, Sood, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Identification and characterization of M6903, an antagonistic anti-TIM-3 monoclonal antibody.
Oncoimmunology, 9, 2020
|
|
7V6J
 
 | | LcCOMT in complex with SAM | | Descriptor: | LcCOMT, S-ADENOSYLMETHIONINE, SODIUM ION | | Authors: | Yu, Y, CHen, Q. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure basis of the caffeic acid O-methyltransferase from Ligusiticum chuanxiong to understand its selective mechanism.
Int.J.Biol.Macromol., 194, 2022
|
|
7V6L
 
 | | LcCOMT in complex with SAH | | Descriptor: | LcCOMT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yu, Y, CHen, Q. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structure basis of the caffeic acid O-methyltransferase from Ligusiticum chuanxiong to understand its selective mechanism.
Int.J.Biol.Macromol., 194, 2022
|
|
8GNA
 
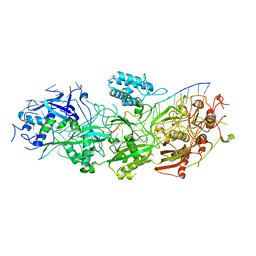 | | Structure of the SbCas7-11-crRNA-NTR complex | | Descriptor: | RAMP superfamily protein, RNA (32-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*U)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
8GU6
 
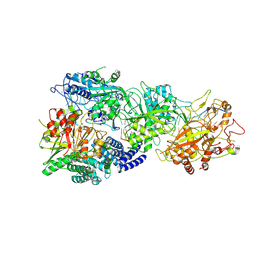 | | Structure of the SbCas7-11-crRNA-NTR-Csx29 complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-09-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
7CND
 
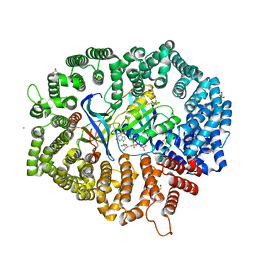 | | NCI-1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-07-31 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of the First Noncovalent Small-Molecule Inhibitor of CRM1.
J.Med.Chem., 64, 2021
|
|
6LQ9
 
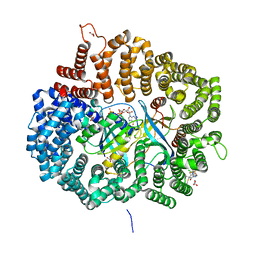 | | S109 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (3~{R},4~{S})-1-[[6-chloranyl-5-(trifluoromethyl)pyridin-2-yl]amino]-3,4-dimethyl-pyrrolidine-2,5-dione, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Design of the First Noncovalent Small-Molecule Inhibitor of CRM1.
J.Med.Chem., 64, 2021
|
|
8J9Z
 
 | |
8J9Y
 
 | |
7X7R
 
 | | Cryo-EM structure of a bacterial protein | | Descriptor: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*AP*GP*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7X8A
 
 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7XC7
 
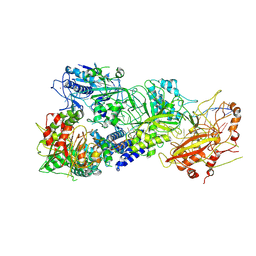 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7X7A
 
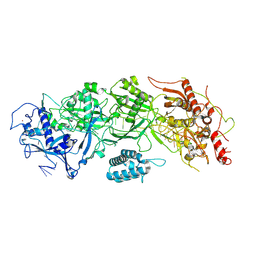 | | Cryo-EM structure of SbCas7-11 in complex with crRNA and target RNA | | Descriptor: | RAMP superfamily protein, RNA (33-MER), ZINC ION | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
