3PZF
 
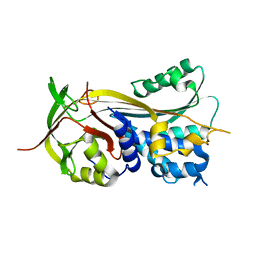 | | 1.75A resolution structure of Serpin-2 from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, An, C, Michel, K. | | Deposit date: | 2010-12-14 | | Release date: | 2011-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of native Anopheles gambiae serpin-2, a negative regulator of melanization in mosquitoes.
Proteins, 79, 2011
|
|
4RSQ
 
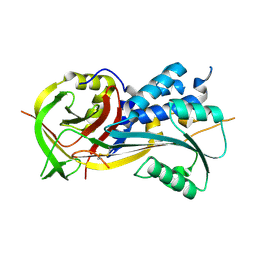 | | 2.9A resolution structure of SRPN2 (K198C/E359C) from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-11-10 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Inhibitory Effects of Hinge Loop Mutagenesis in Serpin-2 from the Malaria Vector Anopheles gambiae.
J.Biol.Chem., 290, 2015
|
|
4ROA
 
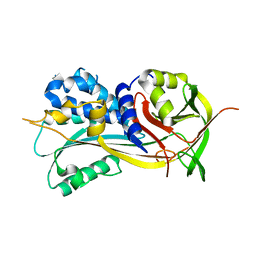 | | 1.90A resolution structure of SRPN2 (S358W) from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Inhibitory Effects of Hinge Loop Mutagenesis in Serpin-2 from the Malaria Vector Anopheles gambiae.
J.Biol.Chem., 290, 2015
|
|
4RO9
 
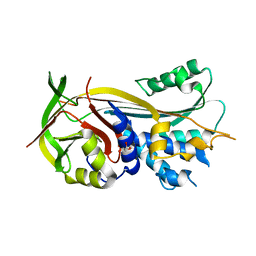 | | 2.0A resolution structure of SRPN2 (S358E) from Anopheles gambiae | | Descriptor: | GLYCEROL, Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Inhibitory Effects of Hinge Loop Mutagenesis in Serpin-2 from the Malaria Vector Anopheles gambiae.
J.Biol.Chem., 290, 2015
|
|
4ROB
 
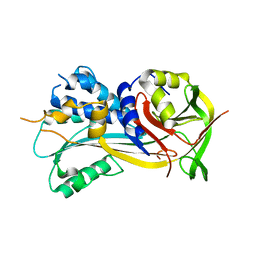 | | 2.8A resolution structure of SRPN2 (K198C) from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8A resolution structure of SRPN2 (K198C) from Anopheles gambiae
To be Published
|
|
1KCQ
 
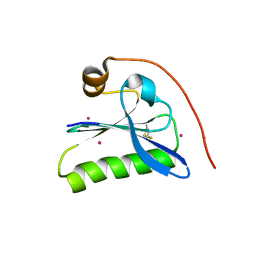 | | Human Gelsolin Domain 2 with a Cd2+ bound | | Descriptor: | CADMIUM ION, GELSOLIN | | Authors: | Kazmirski, S.L, Isaacson, R.L, An, C, Buckle, A, Johnson, C.M, Daggett, V, Fersht, A.R. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Loss of a metal-binding site in gelsolin leads to familial amyloidosis-Finnish type.
Nat.Struct.Biol., 9, 2002
|
|
7REP
 
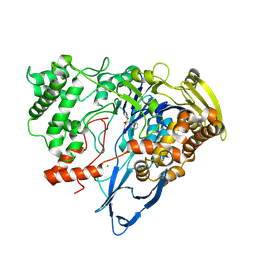 | |
7REO
 
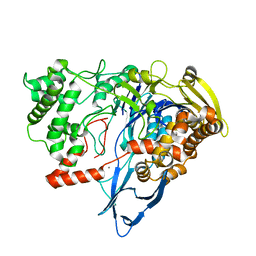 | |
7XR5
 
 | | Crystal structure of imine reductase with NAPDH from Streptomyces albidoflavus | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 6-phosphogluconate dehydrogenase NAD-binding, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Chen, R.C, Gao, S.S. | | Deposit date: | 2022-05-09 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Actinomycetes-derived imine reductases with a preference towards bulky amine substrates.
Commun Chem, 5, 2022
|
|
7XE8
 
 | |
7MHC
 
 | | Structure of human STING in complex with MK-1454 | | Descriptor: | (2R,5R,7R,8S,10R,12aR,14R,15S,15aR,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-15,16-difluoro-2,10-bis(sulfanyl)octahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A kinase-cGAS cascade to synthesize a therapeutic STING activator.
Nature, 603, 2022
|
|
8XUV
 
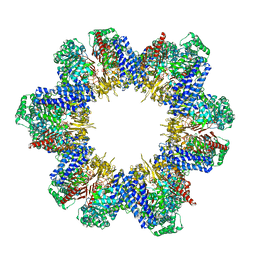 | | Cryo-EM structure of tomato NRC2 filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
8XUO
 
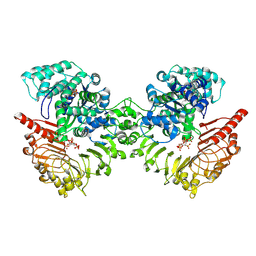 | | Cryo-EM structure of tomato NRC2 dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-13 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
8XUQ
 
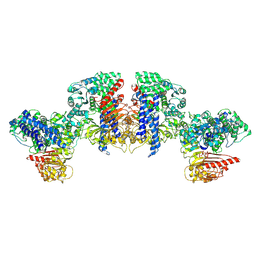 | | Cryo-EM structure of tomato NRC2 tetramer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
5HAD
 
 | |
5GTB
 
 | |
