8E37
 
 | | Structure of Campylobacter concisus wild-type SeMet PglC | | Descriptor: | N,N'-diacetylbacilliosaminyl-1-phosphate transferase | | Authors: | Vuksanovic, N, Ray, L.C, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-08-16 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Synergistic computational and experimental studies of a phosphoglycosyl transferase membrane/ligand ensemble.
J.Biol.Chem., 299, 2023
|
|
8EHV
 
 | |
8EJR
 
 | |
8EJS
 
 | |
8G1N
 
 | | Structure of Campylobacter concisus PglC I57M/Q175M Variant with modeled C-terminus | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, MAGNESIUM ION, N,N'-diacetylbacilliosaminyl-1-phosphate transferase, ... | | Authors: | Dodge, G.J, Ray, L.C, Das, D, Imperiali, B, Allen, K.N. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Co-conserved sequence motifs are predictive of substrate specificity in a family of monotopic phosphoglycosyl transferases.
Protein Sci., 32, 2023
|
|
2YBD
 
 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 with bound phosphate | | Descriptor: | HYDROLASE, HALOACID DEHALOGENASE-LIKE FAMILY, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable Had Family Hydrolase from Pseudomonas Fluorescens Pf-5 with Bound Phosphate
To be Published
|
|
1U7O
 
 | | Magnesium Dependent Phosphatase 1 (MDP-1) | | Descriptor: | ACETATE ION, magnesium-dependent phosphatase-1 | | Authors: | Peisach, E, Selengut, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of the hypothetical phosphotyrosine phosphatase MDP-1 of the haloacid dehalogenase superfamily
Biochemistry, 43, 2004
|
|
1U7P
 
 | | X-ray Crystal Structure of the Hypothetical Phosphotyrosine Phosphatase MDP-1 of the Haloacid Dehalogenase Superfamily | | Descriptor: | MAGNESIUM ION, TUNGSTATE(VI)ION, magnesium-dependent phosphatase-1 | | Authors: | Peisach, E, Selengut, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Crystal Structure of the Hypothetical Phosphotyrosine
Phosphatase MDP-1 of the Haloacid Dehalogenase Superfamily
Biochemistry, 43, 2004
|
|
6P2D
 
 | |
6ALD
 
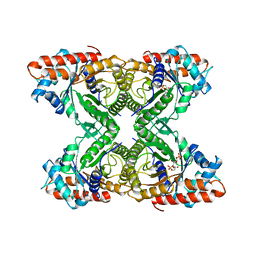 | | RABBIT MUSCLE ALDOLASE A/FRUCTOSE-1,6-BISPHOSPHATE COMPLEX | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), FRUCTOSE-1,6-BIS(PHOSPHATE) ALDOLASE | | Authors: | Choi, K.H, Mazurkie, A.S, Morris, A.J, Utheza, D, Tolan, D.R, Allen, K.N. | | Deposit date: | 1998-12-23 | | Release date: | 2000-01-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a fructose-1,6-bis(phosphate) aldolase liganded to its natural substrate in a cleavage-defective mutant at 2.3 A(,).
Biochemistry, 38, 1999
|
|
4DCC
 
 | | Crystal structure of had family enzyme bt-2542 from bacteroides thetaiotaomicron (target efi-501088) | | Descriptor: | CHLORIDE ION, Putative haloacid dehalogenase-like hydrolase, SODIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
4DWO
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Vetting, M.W, Wasserman, S.R, Morisco, L.L, Sojitra, S, Allen, K.N, Dunaway-Mariano, D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-26 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II
To be Published
|
|
4E38
 
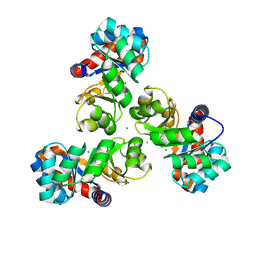 | | Crystal structure of probable keto-hydroxyglutarate-aldolase from Vibrionales bacterium SWAT-3 (Target EFI-502156) | | Descriptor: | CHLORIDE ION, Keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate aldolase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-09 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of probable keto-hydroxyglutarate-aldolase from Vibrionales bacterium SWAT-3 (Target EFI-502156)
To be Published
|
|
4DW8
 
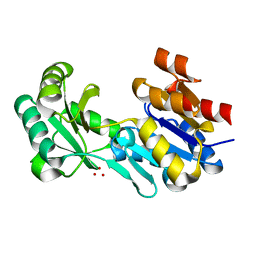 | | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Na crystal form I | | Descriptor: | Haloacid dehalogenase-like hydrolase, SODIUM ION, UNKNOWN LIGAND | | Authors: | Vetting, M.W, Wasserman, S.R, Morisco, L.L, Sojitra, S, Allen, K.N, Dunaway-Mariano, D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-24 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Na crystal form I
To be Published
|
|
6C71
 
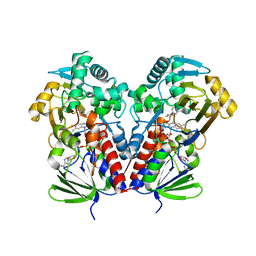 | | Nicotine Oxidoreductase in Complex with S-nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tararina, M.A, Allen, K.N. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Crystallography Coupled with Kinetic Analysis Provides Mechanistic Underpinnings of a Nicotine-Degrading Enzyme.
Biochemistry, 57, 2018
|
|
6CFS
 
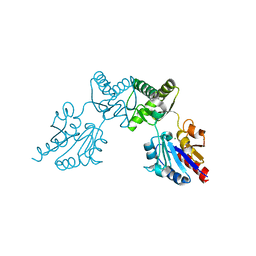 | |
6CFV
 
 | |
4EEK
 
 | | Crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) With bound phosphate and sodium | | Descriptor: | Beta-phosphoglucomutase-related protein, PHOSPHATE ION, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
6CFT
 
 | |
4EEL
 
 | | Crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) with bound citrate and sodium | | Descriptor: | Beta-phosphoglucomutase-related protein, CITRIC ACID, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
4EEN
 
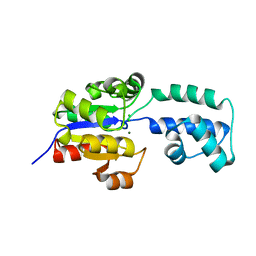 | | crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) with bound magnesium | | Descriptor: | Beta-phosphoglucomutase-related protein, CHLORIDE ION, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
6CFR
 
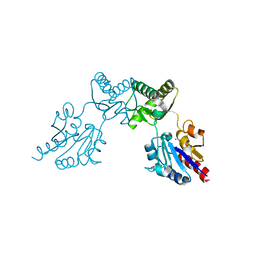 | |
6CFU
 
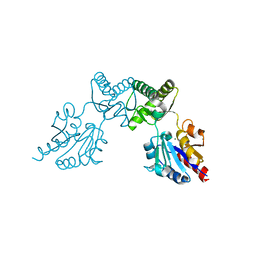 | |
2OJR
 
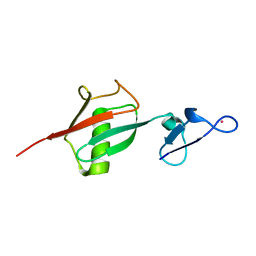 | |
2RB5
 
 | |
