2YMJ
 
 | |
3M4X
 
 | |
6EX9
 
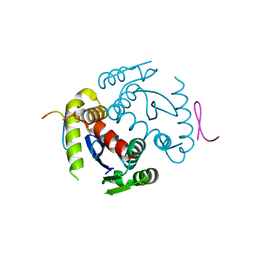 | |
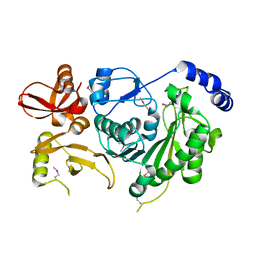
5EU7
 
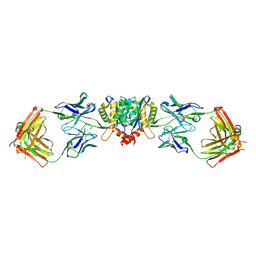 | | Crystal structure of HIV-1 integrase catalytic core in complex with Fab | | Descriptor: | FAB Heavy Chain, FAB light chain, Integrase | | Authors: | Galilee, M, Griner, S.L, Stroud, R.M, Alian, A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Preserved HTH-Docking Cleft of HIV-1 Integrase Is Functionally Critical.
Structure, 24, 2016
|
|
4INK
 
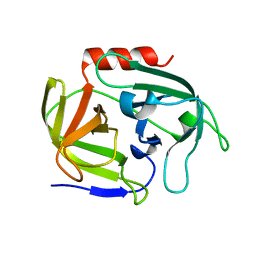 | | Crystal structure of SplD protease from Staphylococcus aureus at 1.56 A resolution | | Descriptor: | Serine protease SplD | | Authors: | Zdzalik, M, Kalinska, M, Cichon, P, Wysocka, M, Stec-Niemczyk, J, Stennicke, H.R, Jabaiah, A, Markiewicz, M, Wladyka, B, Daugherty, P.S, Lesner, A, Rolka, K, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2013-01-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Biochemical and Structural Characterization of SplD Protease from Staphylococcus aureus.
Plos One, 8, 2013
|
|
2J28
 
 | | MODEL OF E. COLI SRP BOUND TO 70S RNCS | | Descriptor: | 23S RIBOSOMAL RNA, 4.5S SIGNAL RECOGNITION PARTICLE RNA, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Halic, M, Blau, M, Becker, T, Mielke, T, Pool, M.R, Wild, K, Sinning, I, Beckmann, R. | | Deposit date: | 2006-08-16 | | Release date: | 2006-11-08 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Following the Signal Sequence from Ribosomal Tunnel Exit to Signal Recognition Particle
Nature, 444, 2006
|
|
2J37
 
 | | MODEL OF MAMMALIAN SRP BOUND TO 80S RNCS | | Descriptor: | 60S RIBOSOMAL PROTEIN L23, RIBOSOMAL PROTEIN L31, RIBOSOMAL PROTEIN L35, ... | | Authors: | Halic, M, Blau, M, Becker, T, Mielke, T, Pool, M.R, Wild, K, Sinning, I, Beckmann, R. | | Deposit date: | 2006-08-18 | | Release date: | 2006-11-08 | | Last modified: | 2019-04-10 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Following the signal sequence from ribosomal tunnel exit to signal recognition particle.
Nature, 444, 2006
|
|
2GO5
 
 | | Structure of signal recognition particle receptor (SR) in complex with signal recognition particle (SRP) and ribosome nascent chain complex | | Descriptor: | SRP RNA, Signal recognition particle 19 kDa protein (SRP19), Signal recognition particle 54 kDa protein (SRP54), ... | | Authors: | Halic, M, Gartmann, M, Schlenker, O, Mielke, T, Pool, M.R, Sinning, I, Beckmann, R. | | Deposit date: | 2006-04-12 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Signal Recognition Particle Receptor Exposes the Ribosomal Translocon Binding Site
Science, 312, 2006
|
|
4PA1
 
 | |
2XN9
 
 | | Crystal structure of the ternary complex between human T cell receptor, staphylococcal enterotoxin H and human major histocompatibility complex class II | | Descriptor: | ENTEROTOXIN H, GLYCEROL, HEMAGGLUTININ, ... | | Authors: | Saline, M, Rodstrom, K.E.J, Fischer, G, Orekhov, V.Y, Karlsson, B.G, Lindkvist-Petersson, K. | | Deposit date: | 2010-07-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of Superantigen Complexed with Tcr and Mhc Reveals Novel Insights Into Superantigenic T Cell Activation.
Nat.Commun., 1, 2010
|
|
2XNA
 
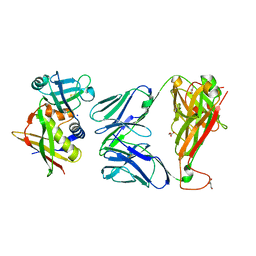 | | Crystal structure of the complex between human T cell receptor and staphylococcal enterotoxin | | Descriptor: | ENTEROTOXIN H, GLYCEROL, SODIUM ION, ... | | Authors: | Saline, M, Rodstrom, K.E.J, Fischer, G, Orekhov, V.Y, Karlsson, B.G, Lindkvist-Petersson, K. | | Deposit date: | 2010-07-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Superantigen Complexed with Tcr and Mhc Reveals Novel Insights Into Superantigenic T Cell Activation.
Nat.Commun., 1, 2010
|
|
6UID
 
 | |
6UIE
 
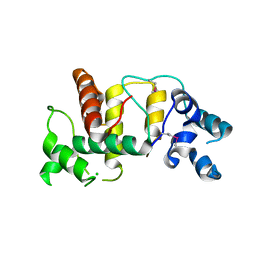 | | Structure of the cytoplasmic domain of the T3SS sorting platform protein PscK from P. aeruginosa | | Descriptor: | CHLORIDE ION, Type III export protein PscK | | Authors: | Muthuramalingam, M, Lovell, S, Battaile, K.P, Picking, W.D. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structures of SctK and SctD from Pseudomonas aeruginosa Reveal the Interface of the Type III Secretion System Basal Body and Sorting Platform.
J.Mol.Biol., 432, 2020
|
|
2X16
 
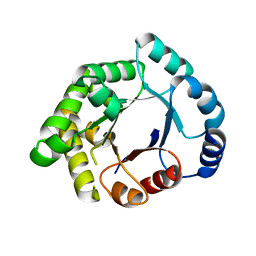 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2009-12-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1T
 
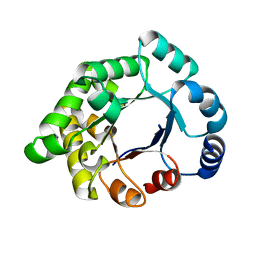 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1R
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 3-(PROPYLSULFONYL)PROPANOIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1U
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X2G
 
 | | CRYSTALLOGRAPHIC BINDING STUDIES WITH AN ENGINEERED MONOMERIC VARIANT OF TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1S
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 3-SULFOPROPANOIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Castejeijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4MQ3
 
 | |
5OVN
 
 | | Crystal Structure of FIV Reverse Transcriptase | | Descriptor: | POL protein | | Authors: | Galilee, M, Alian, A. | | Deposit date: | 2017-08-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.942 Å) | | Cite: | The structure of FIV reverse transcriptase and its implications for non-nucleoside inhibitor resistance.
PLoS Pathog., 14, 2018
|
|
1YSH
 
 | | Localization and dynamic behavior of ribosomal protein L30e | | Descriptor: | 40S RIBOSOMAL PROTEIN S13, RNA (101-MER), RNA (28-MER), ... | | Authors: | Halic, M, Becker, T, Frank, J, Spahn, C.M, Beckmann, R. | | Deposit date: | 2005-02-08 | | Release date: | 2005-07-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Localization and dynamic behavior of ribosomal protein L30e
Nat.Struct.Mol.Biol., 12, 2005
|
|
1RY1
 
 | | Structure of the signal recognition particle interacting with the elongation-arrested ribosome | | Descriptor: | SRP Alu domain, SRP RNA, SRP S domain, ... | | Authors: | Halic, M, Becker, T, Pool, M.R, Spahn, C.M, Grassucci, R.A, Frank, J, Beckmann, R. | | Deposit date: | 2003-12-19 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the signal recognition particle interacting with the elongation-arrested ribosome
Nature, 427, 2004
|
|
2AWV
 
 | | NMR Structural Analysis of the dimer of 5MCCTCATCC | | Descriptor: | 5'-D(*(MCY)P*CP*TP*CP*AP*CP*TP*CP*C)-3' | | Authors: | Canalia, M, Leroy, J.-L. | | Deposit date: | 2005-09-02 | | Release date: | 2005-09-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure, internal motions and association-dissociation kinetics of the i-motif dimer of d(5mCCTCACTCC).
Nucleic Acids Res., 33, 2005
|
|
2N4I
 
 | | The solution structure of Skint-1, a critical determinant of dendritic epidermal gamma-delta T cell selection | | Descriptor: | Selection and upkeep of intraepithelial T-cells protein 1 | | Authors: | Salim, M, Knowles, T.J, Hart, R, Mohammed, F, Woodward, M.J, Willcox, C.R, Overduin, M, Hayday, A.C, Willcox, B.E. | | Deposit date: | 2015-06-18 | | Release date: | 2016-03-02 | | Last modified: | 2016-05-11 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Putative Receptor Binding Surface on Skint-1, a Critical Determinant of Dendritic Epidermal T Cell Selection.
J.Biol.Chem., 291, 2016
|
|
