7E4U
 
 | |
8EX5
 
 | |
8EX6
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1*) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8EX7
 
 | |
8EX4
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8EX8
 
 | |
3OTX
 
 | | Crystal Structure of Trypanosoma brucei rhodesiense Adenosine Kinase Complexed with Inhibitor AP5A | | Descriptor: | Adenosine kinase, putative, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Kuettel, S, Greenwald, J, Kostrewa, D, Ahmed, S, Scapozza, L, Perozzo, R. | | Deposit date: | 2010-09-14 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structures of T. b. rhodesiense Adenosine Kinase Complexed with Inhibitor and Activator: Implications for Catalysis and Hyperactivation
Plos Negl Trop Dis, 5, 2011
|
|
8VKP
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKK
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKM
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKO
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKN
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKL
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
5A8D
 
 | | The high resolution structure of a novel alpha-L-arabinofuranosidase (CtGH43) from Clostridium thermocellum ATCC 27405 | | Descriptor: | ACETATE ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Goyal, A, Ahmed, S, Sharma, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-07-14 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular determinants of substrate specificity revealed by the structure of Clostridium thermocellum arabinofuranosidase 43A from glycosyl hydrolase family 43 subfamily 16.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5A8C
 
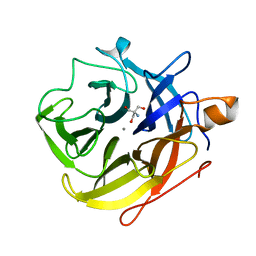 | | The ultra high resolution structure of a novel alpha-L-arabinofuranosidase (CtGH43) from Clostridium thermocellum ATCC 27405 with bound trimethyl N-Oxide (TRS) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6 | | Authors: | Goyal, A, Ahmed, S, Sharma, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-07-14 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Molecular determinants of substrate specificity revealed by the structure of Clostridium thermocellum arabinofuranosidase 43A from glycosyl hydrolase family 43 subfamily 16.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2M02
 
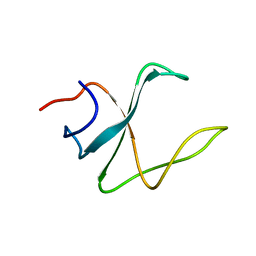 | | 3D structure of cap-gly domain of mammalian dynactin determined by magic angle spinning NMR spectroscopy | | Descriptor: | Dynactin subunit 1 | | Authors: | Yan, S, Hou, G, Schwieters, C.D, Ahmed, S, Williams, J.C, Polenova, T. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Three-Dimensional Structure of CAP-Gly Domain of Mammalian Dynactin Determined by Magic Angle Spinning NMR Spectroscopy: Conformational Plasticity and Interactions with End-Binding Protein EB1.
J.Mol.Biol., 425, 2013
|
|
2OP7
 
 | | WW4 | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP1 | | Authors: | Qin, H.N, Li, M.F, Pu, H, Sankaran, S, Ahmed, S, Song, J.X. | | Deposit date: | 2007-01-27 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the forth WW domain of WWP1
To be Published
|
|
2XTB
 
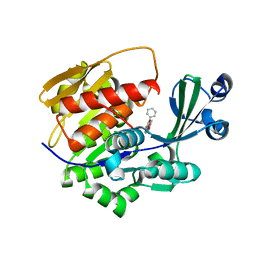 | | Crystal Structure of Trypanosoma brucei rhodesiense Adenosine Kinase Complexed with Activator | | Descriptor: | 4-[5-(4-PHENOXYPHENYL)-1H-PYRAZOL-3-YL]MORPHOLINE, ADENOSINE KINASE | | Authors: | Kuettel, S, Greenwald, J, Kostrewa, D, Ahmed, S, Scapozza, L, Perozzo, R. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of T. B. Rhodesiense Adenosine Kinase Complexed with Inhibitor and Activator: Implications for Catalysis and Hyperactivation.
Plos Negl Trop Dis, 5, 2011
|
|
5V18
 
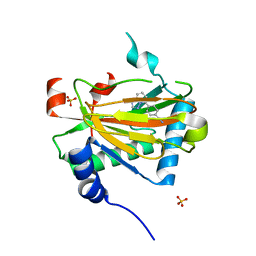 | | Structure of PHD2 in complex with 1,2,4-Triazolo-[1,5-a]pyridine | | Descriptor: | 4-([1,2,4]triazolo[1,5-a]pyridin-5-yl)benzonitrile, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 1,2,4-Triazolo-[1,5-a]pyridine HIF Prolylhydroxylase Domain-1 (PHD-1) Inhibitors With a Novel Monodentate Binding Interaction.
J. Med. Chem., 60, 2017
|
|
5V1B
 
 | | Structure of PHD1 in complex with 1,2,4-Triazolo-[1,5-a]pyridine | | Descriptor: | 4-([1,2,4]triazolo[1,5-a]pyridin-5-yl)benzonitrile, Egl nine homolog 2, FE (III) ION, ... | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | 1,2,4-Triazolo-[1,5-a]pyridine HIF Prolylhydroxylase Domain-1 (PHD-1) Inhibitors With a Novel Monodentate Binding Interaction.
J. Med. Chem., 60, 2017
|
|
1TTQ
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) IN THE PRESENCE OF POTASSIUM AT ROOM TEMPERATURE | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE | | Authors: | Rhee, S, Parris, K, Ahmed, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
Biochemistry, 35, 1996
|
|
1TTP
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) IN THE PRESENCE OF CESIUM, ROOM TEMPERATURE | | Descriptor: | CESIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE | | Authors: | Rhee, S, Parris, K, Ahmed, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
Biochemistry, 35, 1996
|
|
7WVL
 
 | |
8RPO
 
 | | BFL1 in complex with a reversible covalent ligand | | Descriptor: | (~{E})-2-cyano-3-(2-dimethylphosphorylphenyl)-~{N}-[[1-[4-(trifluoromethyl)phenyl]cyclopropyl]methyl]prop-2-enamide, Bcl-2-related protein A1 | | Authors: | Hargreaves, D. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Identification and Evaluation of Reversible Covalent Binders to Cys55 of Bfl-1 from a DNA-Encoded Chemical Library Screen.
Acs Med.Chem.Lett., 15, 2024
|
|
7F8P
 
 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) with new carbapenem drug T203 | | Descriptor: | (2R,3R)-3-methyl-4-(2-oxidanylidene-2-propan-2-yloxy-ethyl)sulfanyl-2-[(2R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]-2,3-dihydro-1H-pyrrole-5-carboxylic acid, (4R,5S,6S)-6-((R)-1-hydroxyethyl)-3-((2-isopropoxy-2-oxoethyl)thio)-4-methyl-7-oxo-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, GLUTAMIC ACID, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2021-07-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric cooperation in beta-lactam binding to a non-classical transpeptidase.
Elife, 11, 2022
|
|
