1H9C
 
 | | NMR structure of cysteinyl-phosphorylated enzyme IIB of the N,N'-diacetylchitobiose specific phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli. | | Descriptor: | PTS SYSTEM, CHITOBIOSE-SPECIFIC IIB COMPONENT | | Authors: | Ab, E, Schuurman-Wolters, G.K, Nijlant, D, Dijkstra, K, Saier, M.H, Robillard, G.T, Scheek, R.M. | | Deposit date: | 2001-03-07 | | Release date: | 2001-05-21 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Cysteinyl-Phosphorylated Enzyme Iib of the N,N'-Diacetylchitobiose Specific Phosphoenolpyruvate-Dependentphosphotransferase System of Escherichia Coli
J.Mol.Biol., 308, 2001
|
|
1E2B
 
 | | NMR STRUCTURE OF THE C10S MUTANT OF ENZYME IIB CELLOBIOSE OF THE PHOSPHOENOL-PYRUVATE DEPENDENT PHOSPHOTRANSFERASE SYSTEM OF ESCHERICHIA COLI, 17 STRUCTURES | | Descriptor: | ENZYME IIB-CELLOBIOSE | | Authors: | Ab, E, Schuurman-Wolters, G, Reizer, J, Saier, M.H, Dijkstra, K, Scheek, R.M, Robillard, G.T. | | Deposit date: | 1996-11-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR side-chain assignments and solution structure of enzyme IIBcellobiose of the phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli.
Protein Sci., 6, 1997
|
|
2CKA
 
 | | Solution structures of the BRK domains of the human Chromo Helicase Domain 7 and 8, reveals structural similarity with GYF domain suggesting a role in protein interaction | | Descriptor: | CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 8 | | Authors: | Ab, E, de Jong, R.N, Diercks, T, Xiaoyun, J, Daniels, M, Kaptein, R, Folkers, G.E. | | Deposit date: | 2006-04-14 | | Release date: | 2007-05-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Brk Domains of the Human Chromo Helicase Domain 7 and 8, Reveals Structural Similarity with Gyf Domain Suggesting a Role in Protein Interaction
To be Published
|
|
2CKC
 
 | | Solution structures of the BRK domains of the human Chromo Helicase Domain 7 and 8, reveals structural similarity with GYF domain suggesting a role in protein interaction | | Descriptor: | CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 7 | | Authors: | Ab, E, de Jong, R.N, Diercks, T, Xiaoyun, J, Daniels, M, Kaptein, R, Folkers, G.E. | | Deposit date: | 2006-04-14 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Brk Domains of the Human Chromo Helicase Domain 7 and 8, Reveals Structural Similarity with Gyf Domain Suggesting a Role in Protein Interaction
To be Published
|
|
2C7H
 
 | | Solution NMR structure of the DWNN domain from human RBBP6 | | Descriptor: | RETINOBLASTOMA-BINDING PROTEIN 6, ISOFORM 3 | | Authors: | Pugh, D.J.R, Ab, E, Faro, A, Lutya, P.T, Hoffmann, E, Rees, D.J.G. | | Deposit date: | 2005-11-24 | | Release date: | 2006-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dwnn, a Novel Ubiquitin-Like Domain, Implicates Rbbp6 in Mrna Processing and Ubiquitin-Like Pathways
Bmc Struct.Biol., 6, 2006
|
|
1W6V
 
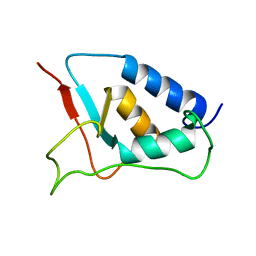 | | Solution structure of the DUSP domain of hUSP15 | | Descriptor: | UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | De Jong, R.D, Ab, E, Diercks, T, Truffault, V, Daniels, M, Kaptein, R, Folkers, G.E. | | Deposit date: | 2004-08-24 | | Release date: | 2006-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Ubiquitin-Specific Protease 15 Dusp Domain.
J.Biol.Chem., 281, 2006
|
|
1Z00
 
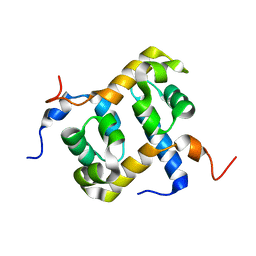 | | Solution structure of the C-terminal domain of ERCC1 complexed with the C-terminal domain of XPF | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Tripsianes, K, Folkers, G, Ab, E, Das, D, Odijk, H, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | Deposit date: | 2005-03-01 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Human ERCC1/XPF Interaction Domains Reveals a Complementary Role for the Two Proteins in Nucleotide Excision Repair
Structure, 13, 2005
|
|
4KLZ
 
 | | Inhibition of Small GTPases by Stabilization of the GDP Complex, a Novel Approach applied to Rit1, a Target for Rheumatoid Arthritis | | Descriptor: | GTP-binding protein Rit1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Shah, D.M, Kobayashi, M, Keizers, P.H, Tuin, A.W, Ab, E, Manning, L, Rzepiela, A.A, Andrews, M, Hoedemaeker, F.J, Siegal, G. | | Deposit date: | 2013-05-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition of Small GTPases by Stabilization of the GDP Complex, a Novel Approach applied to Rit1, a Target for Rheumatoid Arthritis
To be Published
|
|
2BZE
 
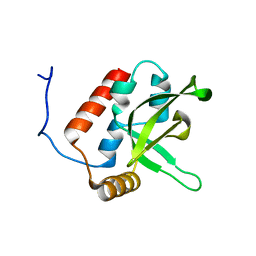 | | NMR Structure of human RTF1 PLUS3 domain. | | Descriptor: | KIAA0252 PROTEIN | | Authors: | Truffault, V, Diercks, T, Ab, E, De Jong, R.N, Daniels, M.A, Kaptein, R, Folkers, G.E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-08-16 | | Release date: | 2007-01-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA Binding of the Human Rtf1 Plus3 Domain.
Structure, 16, 2008
|
|
2C0S
 
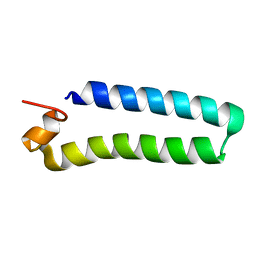 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-09-07 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2BZB
 
 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-08-14 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2BW2
 
 | | BofC from Bacillus subtilis | | Descriptor: | BYPASS OF FORESPORE C | | Authors: | Patterson, H.M, Brannigan, J.A, Cutting, S.M, Wilson, K.S, Wilkinson, A.J, Ab, E, Diercks, T, Folkers, G.E, de Jong, R.N, Truffault, V, Kaptein, R. | | Deposit date: | 2005-07-08 | | Release date: | 2005-09-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of bypass of forespore C, an intercompartmental signaling factor during sporulation in Bacillus.
J. Biol. Chem., 280, 2005
|
|
3ZTG
 
 | | Solution structure of the RING finger-like domain of Retinoblastoma Binding Protein-6 (RBBP6) | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE RBBP6, ZINC ION | | Authors: | Kappo, M.A, Ab, E, Atkinson, R.A, Faro, A, Muleya, V, Mulaudzi, T, Poole, J.O, McKenzie, J.M, Pugh, D.J.R. | | Deposit date: | 2011-07-07 | | Release date: | 2011-12-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ring Finger-Like Domain of Retinoblastoma Binding Protein-6 (Rbbp6) Suggests It Functions as a U-Box
J.Biol.Chem., 287, 2012
|
|
2JM8
 
 | | R21A Spc-SH3 free | | Descriptor: | Spectrin alpha chain, brain | | Authors: | van Nuland, N.A.J, Casares, S, Ab, E, Eshuis, H, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-10-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of the R21A Spc-SH3:P41 complex: Understanding the determinants of binding affinity by comparison with Abl-SH3
Bmc Struct.Biol., 7, 2007
|
|
2JTE
 
 | | Third SH3 domain of CD2AP | | Descriptor: | CD2-associated protein | | Authors: | van Nuland, N.A.J, Ortega, R.J, Romero Romero, M, Ab, E, Ora, A, Lopez, M.O, Azuaga, A.I. | | Deposit date: | 2007-07-28 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The high resolution NMR structure of the third SH3 domain of CD2AP.
J.Biomol.Nmr, 39, 2007
|
|
2JMA
 
 | | R21A Spc-SH3:P41 complex | | Descriptor: | P41 peptide, Spectrin alpha chain, brain | | Authors: | van Nuland, N.A.J, Casares, S, Ab, E, Eshuis, H, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-10-25 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of the R21A Spc-SH3:P41 complex: Understanding the determinants of binding affinity by comparison with Abl-SH3
Bmc Struct.Biol., 7, 2007
|
|
2JM9
 
 | | R21A Spc-SH3 bound | | Descriptor: | Spectrin alpha chain, brain | | Authors: | van Nuland, N.A.J, Casares, S, Ab, E, Eshuis, H, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-10-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of the R21A Spc-SH3:P41 complex: Understanding the determinants of binding affinity by comparison with Abl-SH3
Bmc Struct.Biol., 7, 2007
|
|
2KJ3
 
 | | High-resolution structure of the HET-s(218-289) prion in its amyloid form obtained by solid-state NMR | | Descriptor: | Small s protein | | Authors: | Van Melckebeke, H, Wasmer, C, Lange, A, AB, E, Loquet, A, Meier, B.H. | | Deposit date: | 2009-05-20 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of HET-s(218-289) Amyloid Fibrils by Solid-State NMR Spectroscopy
J.Am.Chem.Soc., 132, 2010
|
|
2K7F
 
 | | HADDOCK calculated model of the complex between the BRCT region of RFC p140 and dsDNA | | Descriptor: | 5'-D(P*DCP*DGP*DAP*DCP*DCP*DTP*DCP*DGP*DAP*DGP*DAP*DTP*DCP*DA)-3', 5'-D(P*DCP*DTP*DCP*DGP*DAP*DGP*DGP*DTP*DCP*DG)-3', Replication factor C subunit 1 | | Authors: | Kobayashi, M, Ab, E, Bonvin, A, Siegal, G. | | Deposit date: | 2008-08-10 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the DNA-bound BRCA1 C-terminal region from human replication factor C p140 and model of the protein-DNA complex.
J.Biol.Chem., 285, 2010
|
|
2GVS
 
 | | NMR solution structure of CSPsg4 | | Descriptor: | chemosensory protein CSP-sg4 | | Authors: | Tomaselli, S, Crescenzi, O, Sanfelice, D, Ab, E, Tancredi, T, Picone, D. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Chemosensory Protein from the Desert Locust Schistocerca gregaria(,).
Biochemistry, 45, 2006
|
|
2K6G
 
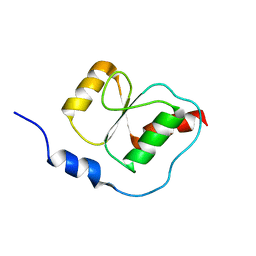 | |
2VKC
 
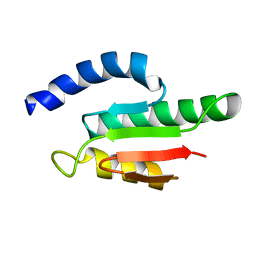 | | Solution structure of the B3BP Smr domain | | Descriptor: | NEDD4-BINDING PROTEIN 2 | | Authors: | Diercks, T, Ab, E, Daniels, M.A, deJong, R.N, Besseling, R, Kaptein, R, Folkers, G.E. | | Deposit date: | 2007-12-18 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the DNA- Binding Activity of the B3BP-Smr Domain.
J.Mol.Biol., 383, 2008
|
|
5P2P
 
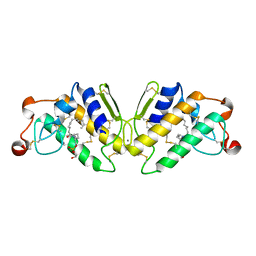 | | X-RAY STRUCTURE OF PHOSPHOLIPASE A2 COMPLEXED WITH A SUBSTRATE-DERIVED INHIBITOR | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2, PHOSPHONIC ACID 2-DODECANOYLAMINO-HEXYL ESTER PROPYL ESTER | | Authors: | Dijkstra, B.W, Thunnissen, M.M.G.M, Kalk, K.H, Drenth, J. | | Deposit date: | 1990-09-01 | | Release date: | 1991-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of phospholipase A2 complexed with a substrate-derived inhibitor.
Nature, 347, 1990
|
|
2WY2
 
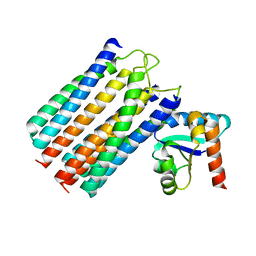 | | NMR structure of the IIAchitobiose-IIBchitobiose phosphoryl transition state complex of the N,N'-diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT, PHOSPHITE ION | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
2WWV
 
 | | NMR structure of the IIAchitobiose-IIBchitobiose complex of the N,N'- diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
