7RU8
 
 | | CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.30 Fab Kappa chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
4WWI
 
 | |
4Y76
 
 | | Factor Xa complex with GTC000401 | | Descriptor: | CALCIUM ION, Coagulation factor X, N~2~-[(6-chloronaphthalen-2-yl)sulfonyl]-N~2~-{(3S)-1-[(2S)-1-(4-methyl-1,4-diazepan-1-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}glycinamide | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
4Y79
 
 | | Factor Xa complex with GTC000406 | | Descriptor: | (E)-2-(4-chlorophenyl)-N-{(3S)-1-[(2S)-1-(morpholin-4-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}ethenesulfonamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
4ZMD
 
 | | C domain of staphylococcal protein A mutant - Q9W | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Deis, L.N, Oas, T.G. | | Deposit date: | 2015-05-03 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Suppression of conformational heterogeneity at a protein-protein interface.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZNC
 
 | |
6A67
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody FLD21.140 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLD21.140 Heavy Chain, FLD21.140 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Complementary recognition of the receptor-binding site of highly pathogenic H5N1 influenza viruses by two human neutralizing antibodies.
J. Biol. Chem., 293, 2018
|
|
2I50
 
 | | Solution Structure of Ubp-M Znf-UBP domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 16, ZINC ION | | Authors: | Pai, M.-T. | | Deposit date: | 2006-08-23 | | Release date: | 2007-08-07 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ubp-M BUZ domain, a highly specific protein module that recognizes the C-terminal tail of free ubiquitin.
J.Mol.Biol., 370, 2007
|
|
3S7L
 
 | |
3S7M
 
 | |
5ZXV
 
 | |
6IUT
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody AVFluIgG01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, AVFluIgG01 Heavy Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional definition of a vulnerable site on the hemagglutinin of highly pathogenic avian influenza A virus H5N1.
J. Biol. Chem., 294, 2019
|
|
6IUV
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody 3C11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3C11 Heavy Chain, 3C11 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | Structural and functional definition of a vulnerable site on the hemagglutinin of highly pathogenic avian influenza A virus H5N1.
J. Biol. Chem., 294, 2019
|
|
2KHU
 
 | | Solution Structure of the Ubiquitin-Binding Motif of Human Polymerase Iota | | Descriptor: | Immunoglobulin G-binding protein G, DNA polymerase iota | | Authors: | Bomar, M.G, D'Souza, S, Bienko, M, Dikic, I, Walker, G. | | Deposit date: | 2009-04-11 | | Release date: | 2010-02-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Unconventional Ubiquitin Recognition by the Ubiquitin-Binding Motif within the Y Family DNA Polymerases iota and Rev1.
Mol.Cell, 37, 2010
|
|
2KHW
 
 | | Solution Structure of the human Polymerase iota UBM2-Ubiquitin Complex | | Descriptor: | Immunoglobulin G-binding protein G, DNA polymerase iota, Ubiquitin | | Authors: | Bomar, M.G, D'Souza, S, Bienko, M, Dikic, I, Walker, G. | | Deposit date: | 2009-04-13 | | Release date: | 2010-02-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the human Polymerase iota UBM2-Ubiquitin Complex
To be Published
|
|
6IEW
 
 | |
6IHJ
 
 | |
4IHF
 
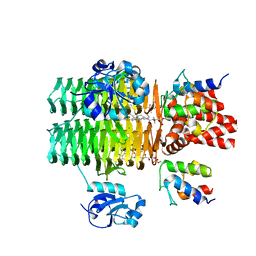 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | Acyl carrier protein, CHLORIDE ION, S-[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] (3R)-3-hydroxytetradecanethioate, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
4IHH
 
 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.132 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
4IHG
 
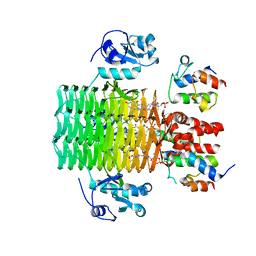 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
8JEC
 
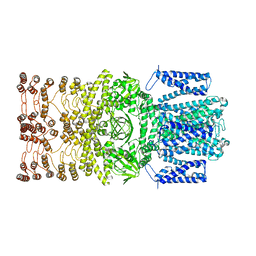 | |
8JEU
 
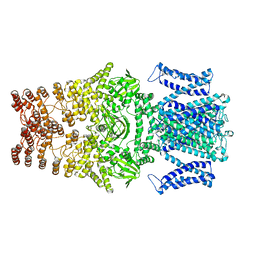 | |
8JET
 
 | |
5XC2
 
 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminarihexaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Qin, Z, Yang, S, Peng, Z, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
1QK6
 
 | | Solution structure of huwentoxin-I by NMR | | Descriptor: | HUWENTOXIN-I | | Authors: | Qu, Y, Liang, S, Ding, J, Liu, X, Zhang, R, Gu, X. | | Deposit date: | 1999-07-10 | | Release date: | 1999-08-20 | | Last modified: | 2019-01-16 | | Method: | SOLUTION NMR | | Cite: | Proton Nuclear Magnetic Resonance Studies on Huwentoxin-I from the Venom of the Spider Selenocosmia Huwena:2.Three-Dimensional Structure in Solution
J.Protein Chem., 16, 1997
|
|
