6T2K
 
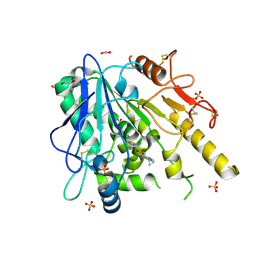 | | Furano[2,3-d]prymidine amides as Notum inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 2-(6-chloranyl-7-cyclopropyl-thieno[3,2-d]pyrimidin-4-yl)sulfanylethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2019-10-08 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Scaffold-hopping identifies furano[2,3-d]pyrimidine amides as potent Notum inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8S7C
 
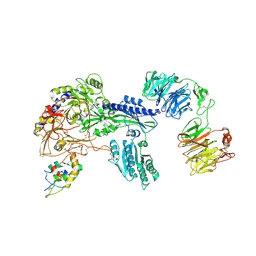 | | Ternary Complex of Cachd1, FZD5 and LRP6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-5, ... | | 著者 | Zhao, Y, Ren, J, Jones, E.Y. | | 登録日 | 2024-02-29 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (4.7 Å) | | 主引用文献 | Cachd1 interacts with Wnt receptors and regulates neuronal asymmetry in the zebrafish brain.
Science, 384, 2024
|
|
7MPJ
 
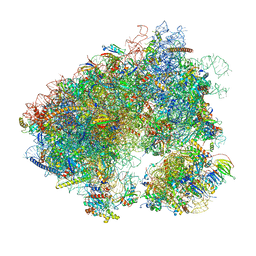 | | Stm1 bound vacant 80S structure isolated from wild-type | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Rai, J, Zhao, Y, Li, H. | | 登録日 | 2021-05-04 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7MPI
 
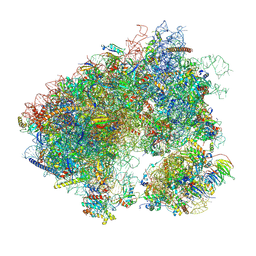 | | Stm1 bound vacant 80S structure isolated from cbf5-D95A | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Rai, J, Zhao, Y, Li, H. | | 登録日 | 2021-05-04 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7N8B
 
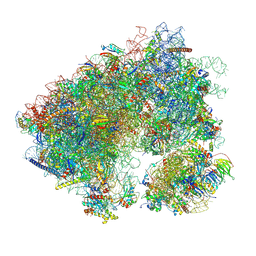 | | Cycloheximide bound vacant 80S structure isolated from cbf5-D95A | | 分子名称: | 18S RIBOSOMAL RNA, 25S, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | 著者 | Rai, J, Zhao, Y, Li, H. | | 登録日 | 2021-06-14 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7TTN
 
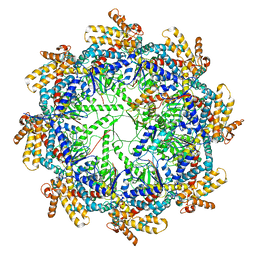 | | The beta-tubulin folding intermediate II | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | 著者 | Zhao, Y, Frydman, J, Chiu, W. | | 登録日 | 2022-02-01 | | 公開日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TRG
 
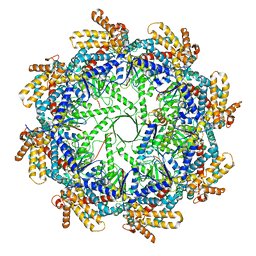 | | The beta-tubulin folding intermediate I | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | 著者 | Zhao, Y, Frydman, J, Chiu, W. | | 登録日 | 2022-01-28 | | 公開日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TUB
 
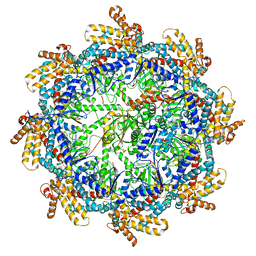 | | The beta-tubulin folding intermediate IV | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | 著者 | Zhao, Y, Frydman, J, Chiu, W. | | 登録日 | 2022-02-02 | | 公開日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TTT
 
 | | The beta-tubulin folding intermediate III | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | 著者 | Zhao, Y, Frydman, J, Chiu, W. | | 登録日 | 2022-02-01 | | 公開日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
5B6G
 
 | | Protein-protein interaction | | 分子名称: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | 著者 | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | 登録日 | 2016-05-27 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | 登録日 | 2020-02-19 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1PTY
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH TWO PHOSPHOTYROSINE MOLECULES | | 分子名称: | MAGNESIUM ION, O-PHOSPHOTYROSINE, PROTEIN TYROSINE PHOSPHATASE 1B | | 著者 | Zhao, Y, Puius, Y.A, Sullivan, M, Lawrence, D, Almo, S.C, Zhang, Z.-Y. | | 登録日 | 1997-01-16 | | 公開日 | 1998-01-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | 著者 | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | 登録日 | 2020-04-08 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5F56
 
 | | Structure of RecJ complexed with DNA and SSB-ct | | 分子名称: | ALA-ASP-LEU-PRO-PHE, DNA (5'-D(*CP*TP*GP*AP*TP*GP*GP*CP*A)-3'), MANGANESE (II) ION, ... | | 著者 | Zhao, Y, Hua, Y, Cheng, K. | | 登録日 | 2015-12-04 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
5IZ8
 
 | | Protein-protein interaction | | 分子名称: | ACE-ALA-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein, TRIETHYLENE GLYCOL | | 著者 | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | 登録日 | 2016-03-25 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZA
 
 | | Protein-protein interaction | | 分子名称: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | 著者 | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | 登録日 | 2016-03-25 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ9
 
 | | Protein-protein interaction | | 分子名称: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein | | 著者 | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | 登録日 | 2016-03-25 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
6LPM
 
 | |
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp6/7 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp9/10 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp4/5 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-14 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp14/15 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp15/16 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E5X
 
 | |
