7T1F
 
 | | Crystal structure of GDP-bound T50I mutant of human KRAS4B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analyses of a germline KRAS T50I mutation provide insights into Raf activation.
JCI Insight, 8, 2023
|
|
8E4V
 
 | | Solution structure of the WH domain of MORF | | Descriptor: | Isoform 3 of Histone acetyltransferase KAT6B | | Authors: | Zhang, Y, Kutateladze, T.G. | | Deposit date: | 2022-08-19 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | MORF and MOZ acetyltransferases target unmethylated CpG islands through the winged helix domain.
Nat Commun, 14, 2023
|
|
5LS6
 
 | | Structure of Human Polycomb Repressive Complex 2 (PRC2) with inhibitor | | Descriptor: | 1-[(1~{R})-1-[1-[2,2-bis(fluoranyl)propyl]piperidin-4-yl]ethyl]-~{N}-[(4-methoxy-6-methyl-2-oxidanylidene-3~{H}-pyridin-3-yl)methyl]-2-methyl-indole-3-carboxamide, Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2, Jarid2 K116me3, ... | | Authors: | Zhang, Y, Justin, N, Chen, S, Wilson, J, Gamblin, S. | | Deposit date: | 2016-08-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas.
J. Med. Chem., 59, 2016
|
|
6C00
 
 | |
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
2Q5A
 
 | | human Pin1 bound to L-PEPTIDE | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE, Five residue peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Noel, J.P, Zhang, Y. | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for high-affinity peptide inhibition of human Pin1.
Acs Chem.Biol., 2, 2007
|
|
2QCX
 
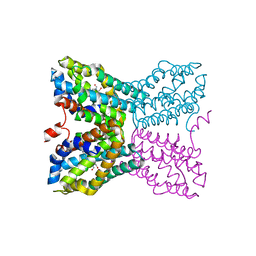 | | Crystal structure of Bacillus subtilis TenA Y112F mutant complexed with formyl aminomethyl pyrimidine | | Descriptor: | N-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-N-(2-HYDROXYETHYL)FORMAMIDE, Transcriptional activator tenA | | Authors: | Zhang, Y, Jenkins, A.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2007-06-19 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis studies on TenA: A thiamin salvage enzyme from Bacillus subtilis
Bioorg.Chem., 36, 2008
|
|
2VI5
 
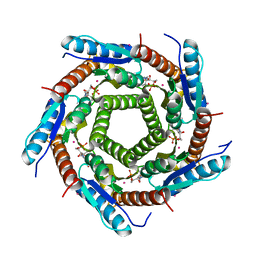 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO N-6-(ribitylamino)pyrimidine-2,4(1H,3H)-dione-5-yl-propionamide | | Descriptor: | 1-deoxy-1-{[(5S)-2,6-dioxo-5-(propanoylamino)-1,2,5,6-tetrahydropyrimidin-4-yl]amino}-D-ribitol, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, PHOSPHATE ION, ... | | Authors: | Morgunova, E, Zhang, Y, Jin, G, Illarionov, B, Bacher, A, Fischer, M, Cushman, M, Ladenstein, R. | | Deposit date: | 2007-11-27 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A New Series of N-[2,4-Dioxo-6-D-Ribitylamino-1,2, 3,4-Tetrahydropyrimidin-5-Yl]Oxalamic Acid Derivatives as Inhibitors of Lumazine Syntase and Riboflavin Synthase: Design, Synthesis, Biochemical Evaluation, Crystallography and Mechanistic Implications.
J.Org.Chem., 73, 2008
|
|
3OG7
 
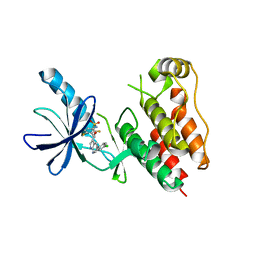 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX4032 | | Descriptor: | AKAP9-BRAF fusion protein, N-(3-{[5-(4-chlorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)propane-1-sulfonamide | | Authors: | Zhang, Y, Zhang, K.Y, Zhang, C. | | Deposit date: | 2010-08-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma.
Nature, 467, 2010
|
|
4HVS
 
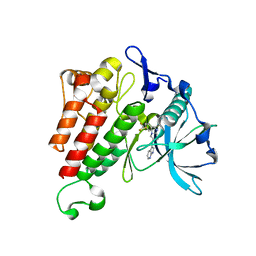 | |
4HW7
 
 | |
5CPE
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-cycloheptyl-1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CP5
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-N-(4-fluorophenyl)-2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5COI
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1-({[(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)sulfonyl]amino}methyl)cyclopentanecarboxylic acid, Bromodomain-containing protein 4 | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CS8
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5D0C
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-08-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CTL
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | Bromodomain-containing protein 4, N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)benzenesulfonamide | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CRM
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CQT
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | Bromodomain-containing protein 4, N-cyclohexyl-1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CY9
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-30 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CRZ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-4-fluorobenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5DX4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-2-methoxybenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
4LA2
 
 | | Crystal structure of dimethylsulphoniopropionate (DMSP) lyase DddQ | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dimethylsulphoniopropionate (DMSP) lyase DddQ, ZINC ION | | Authors: | Zhang, Y, Li, C. | | Deposit date: | 2013-06-19 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insight into bacterial cleavage of oceanic dimethylsulfoniopropionate into dimethyl sulfide
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LA3
 
 | |
5E17
 
 | |
