8K8J
 
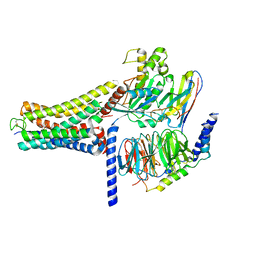 | | Cannabinoid Receptor 1 bound to Fenofibrate coupling MiniGsq and Nb35 Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Tang, W.Q, Wang, T.X, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Fenofibrate Recognition and G q Protein Coupling Mechanisms of the Human Cannabinoid Receptor CB1.
Adv Sci, 11, 2024
|
|
4Y9L
 
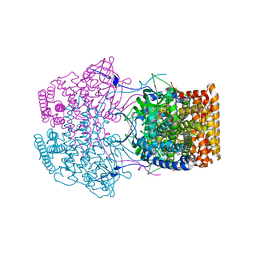 | | Crystal Structure of Caenorhabditis elegans ACDH-11 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Protein ACDH-11, isoform b | | Authors: | Li, Z.J, Zhai, Y.J, Zhang, K, Sun, F. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Acyl-CoA Dehydrogenase Drives Heat Adaptation by Sequestering Fatty Acids
Cell, 161, 2015
|
|
5X9B
 
 | |
5ZU5
 
 | |
6AVO
 
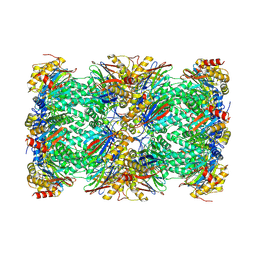 | | Cryo-EM structure of human immunoproteasome with a novel noncompetitive inhibitor that selectively inhibits activated lymphocytes | | Descriptor: | N~1~-{2-[([1,1'-biphenyl]-3-carbonyl)amino]ethyl}-N~4~-tert-butyl-N~2~-(3-phenylpropanoyl)-L-aspartamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Li, H, Santos, R, Bai, L. | | Deposit date: | 2017-09-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of human immunoproteasome with a reversible and noncompetitive inhibitor that selectively inhibits activated lymphocytes.
Nat Commun, 8, 2017
|
|
7EUY
 
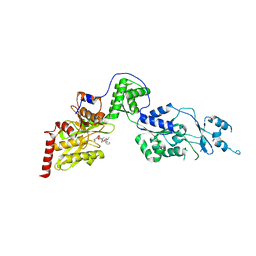 | | Crystal structure of the Lon-like protease MtaLonC with D582A mutation in complex with substrate polypeptide | | Descriptor: | ALA-PRO-GLU-ALA-VAL, Endopeptidase La, PHOSPHATE ION | | Authors: | Hsieh, K.Y, Kuo, C.I, Su, S.C, Huang, K.F, Chang, C.I. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7EV4
 
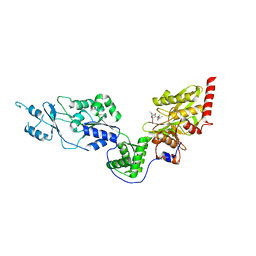 | | Crystal structure of the Lon-like protease MtaLonC with S582A mutation in complex with F-b20-Q | | Descriptor: | Endopeptidase La, F-b20-Q peptide {ortho-aminobenzoic acid (Abz)- QLRSLNGEWRFAWFPAPEAV[Tyr(3-NO2)]A}, PHOSPHATE ION | | Authors: | Hsieh, K.Y, Kuo, C.I, Su, S.C, Huang, K.F, Chang, C.I. | | Deposit date: | 2021-05-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7EUX
 
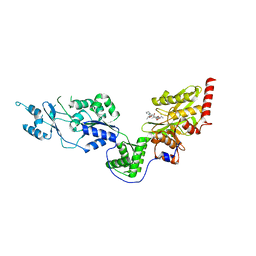 | | Crystal structure of the Lon-like protease MtaLonC with D581A mutation in complex with substrate polypeptide | | Descriptor: | ALA-PRO-GLU-ALA-VAL, Endopeptidase La, PHOSPHATE ION | | Authors: | Hsieh, K.Y, Kuo, C.I, Su, S.C, Huang, K.F, Chang, C.I. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7EV6
 
 | | Crystal structure of the Lon-like protease MtaLonC with D581A mutation in complex with F-b20-Q | | Descriptor: | Endopeptidase La, F-b20-Q peptide {ortho-aminobenzoic acid (Abz)- QLRSLNGEWRFAWFPAPEAV[Tyr(3-NO2)]A}, PHOSPHATE ION | | Authors: | Hsieh, K.Y, Kuo, C.I, Su, S.C, Huang, K.F, Chang, C.I. | | Deposit date: | 2021-05-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7XA9
 
 | | Structure of Arabidopsis thaliana CLCa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Ji, S, Jin, H, Kaiming, Z, Mingxing, W, Shanshan, L, Long, C. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the plant nitrate transporter AtCLCa reveals characteristics of the anion-binding site and the ATP-binding pocket.
J.Biol.Chem., 299, 2023
|
|
7Y38
 
 | |
7YPM
 
 | | Crystal structure of transaminase CC1012 complexed with PLP and L-alanine | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aspartate aminotransferase family protein, ... | | Authors: | Yang, L, Wang, H, Wei, D. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Mechanism-Guided Computational Design of omega-Transaminase by Reprograming of High-Energy-Barrier Steps.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7YPN
 
 | | Crystal structure of transaminase CC1012 mutant M9 complexed with PLP | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase family protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, L, Wang, H, Wei, D. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism-Guided Computational Design of omega-Transaminase by Reprograming of High-Energy-Barrier Steps.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y18
 
 | |
7Y17
 
 | |
7Y16
 
 | | Crystal structure of rRNA-processing protein Las1 | | Descriptor: | LAS1 protein | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2022-06-07 | | Release date: | 2023-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
7K58
 
 | |
6ITG
 
 | | a new alginate lyase (PL6) from Vibrio splendidus OU02 | | Descriptor: | Alginate lyase, GLYCEROL, MALONATE ION | | Authors: | Liu, W.Z, Lyu, Q.Q. | | Deposit date: | 2018-11-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural insights into a novel Ca2+-independent PL-6 alginate lyase from Vibrio OU02 identify the possible subsites responsible for product distribution.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
