2VBS
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with PO4 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, RIBOFLAVIN KINASE, ... | | Authors: | Hartmann, M.D, Djuranovic, S, Ammelburg, M, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBV
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and FMN | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBT
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and PO4 | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, PHOSPHATE ION, RIBOFLAVIN KINASE, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2W9R
 
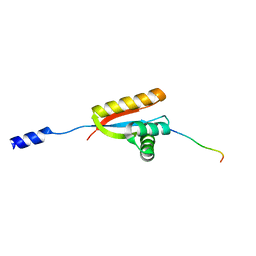 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, DNA PROTECTION DURING STARVATION PROTEIN | | Authors: | Schuenemann, V, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-01-28 | | Release date: | 2009-04-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
2W1T
 
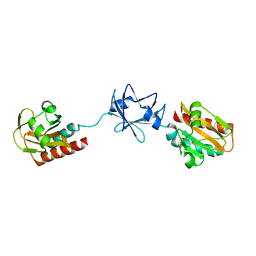 | | Crystal Structure of B. subtilis SpoVT | | Descriptor: | STAGE V SPORULATION PROTEIN T | | Authors: | Asen, I, Djuranovic, S, Lupas, A.N, Zeth, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-18 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Spovt, the Final Modulator of Gene Expression During Spore Development in Bacillus Subtilis
J.Mol.Biol., 386, 2009
|
|
2WA8
 
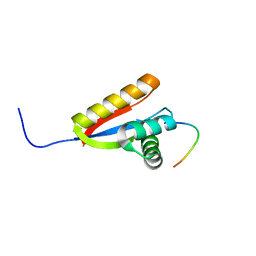 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS - The Phe peptide structure | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, N-END RULE PEPTIDE | | Authors: | Schuenemann, V.J, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
2W1R
 
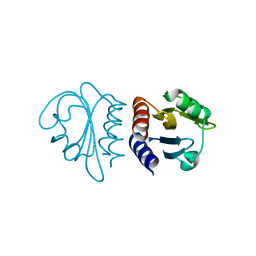 | | Crystal Structure of the C-terminal Domain of B. subtilis SpoVT | | Descriptor: | STAGE V SPORULATION PROTEIN T | | Authors: | Asen, I, Djuranovic, S, Lupas, A.N, Zeth, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-18 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Spovt, the Final Modulator of Gene Expression During Spore Development in Bacillus Subtilis
J.Mol.Biol., 386, 2009
|
|
2WA9
 
 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS - Trp peptide structure | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, TRP PEPTIDE | | Authors: | Schuenemann, V.J, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-28 | | Last modified: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
2WFW
 
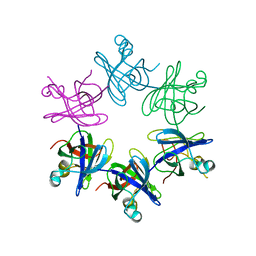 | | Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases - The Arc domain structure | | Descriptor: | ARC | | Authors: | Djuranovic, S, Hartmann, M.D, Habeck, M, Ursinus, A, Zwickl, P, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2WG5
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4 | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2WG6
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4, P61A Mutant | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2X8X
 
 | |
2XTR
 
 | | Structure of the P176A Colicin M mutant from E. coli | | Descriptor: | COLICIN-M, NITRATE ION | | Authors: | Helbig, S, Patzer, S.I, Braun, V, Zeth, K. | | Deposit date: | 2010-10-12 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Activation of Colicin M by the Fkpa Prolyl Cis- Trans Isomerase/Chaperone.
J.Biol.Chem., 286, 2011
|
|
2XTQ
 
 | |
2YH3
 
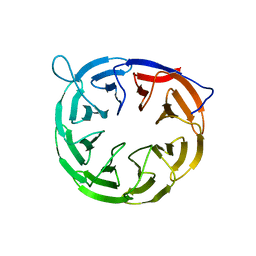 | | The structure of BamB from E. coli | | Descriptor: | LIPOPROTEIN YFGL | | Authors: | Albrecht, R, Zeth, K. | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-25 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Outer Membrane Protein Biogenesis in Bacteria.
J.Biol.Chem., 286, 2011
|
|
4GDO
 
 | | Structure of a fragment of the rod domain of plectin | | Descriptor: | Plectin | | Authors: | De Pereda, J.M, Buey, R.M, Uson, I, Sammito, M.D, De Marino, I. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploiting tertiary structure through local folds for crystallographic phasing.
Nat.Methods, 10, 2013
|
|
2L7I
 
 | |
4RQY
 
 | |
2L7H
 
 | |
6V06
 
 | | Crystal structure of Beta-2 glycoprotein I purified from plasma (pB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-glycoprotein 1, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
6V09
 
 | | Crystal structure of human recombinant Beta-2 glycoprotein I short tag (ST-B2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
6V08
 
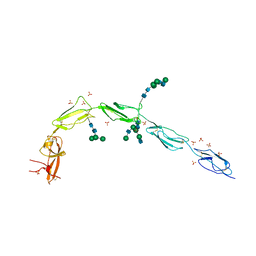 | | Crystal structure of human recombinant Beta-2 glycoprotein I (hrB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
2LFR
 
 | | Solution structure of the chimeric Af1503 HAMP- EnvZ DHp homodimer | | Descriptor: | HAMP domain-containing protein, Osmolarity sensor protein EnzV chimera | | Authors: | Coles, M, Ferris, H.U, Hulko, M, Martin, J, Lupas, A.N. | | Deposit date: | 2011-07-10 | | Release date: | 2011-08-24 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | Mechanism of regulation of receptor histidine kinases.
Structure, 20, 2012
|
|
2M7T
 
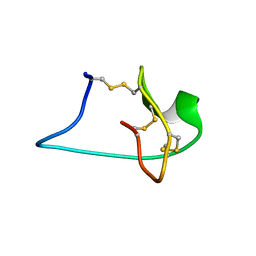 | | Solution NMR Structure of Engineered Cystine Knot Protein 2.5D | | Descriptor: | Cystine Knot Protein 2.5D | | Authors: | Cochran, F.V, Das, R. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
2P3M
 
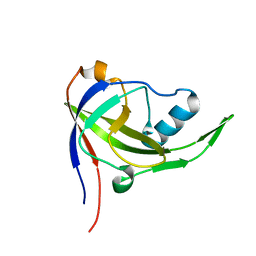 | | Solution structure of Mj0056 | | Descriptor: | Riboflavin Kinase MJ0056 | | Authors: | Coles, M, Truffault, V, Djuranovic, S, Martin, J, Lupas, A.N. | | Deposit date: | 2007-03-09 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A CTP-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
