4MJ5
 
 | | Crystal Structure of HLA-A*1101 in complex with H1-22, an influenza A(H1N1) virus epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Liu, J, Tan, S, Zhao, M, Qi, J, Gao, G.F. | | Deposit date: | 2013-09-03 | | Release date: | 2014-10-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Cross-immunity Against Avian Influenza A(H7N9) Virus in the Healthy Population Is Affected by Antigenicity-Dependent Substitutions.
J.Infect.Dis., 214, 2016
|
|
4MJ6
 
 | | Crystal Structure of HLA-A*1101 in complex with H7-22, an influenza A(H7N9) virus epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Liu, J, Tan, S, Zhao, M, Qi, J, Gao, G.F. | | Deposit date: | 2013-09-03 | | Release date: | 2014-10-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Cross-immunity Against Avian Influenza A(H7N9) Virus in the Healthy Population Is Affected by Antigenicity-Dependent Substitutions.
J.Infect.Dis., 214, 2016
|
|
7ENE
 
 | |
7EN9
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-02 | | Descriptor: | 3C-like proteinase, 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7EN8
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-04 | | Descriptor: | 3C-like proteinase, GLYCEROL, ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7END
 
 | |
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | Descriptor: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6D
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | Descriptor: | Infectious bronchitis virus (IBV) main protease | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6G
 
 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | Descriptor: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2EX1
 
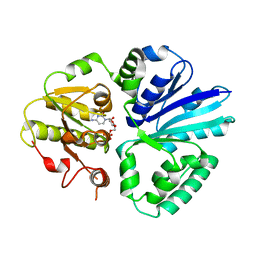 | | Crystal structure of mutifunctional sialyltransferase from Pasteurella multocida with CMP bound | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, a2,3-sialyltransferase, a2,a6-sialyltransferase | | Authors: | Ni, L, Sun, M, Chen, X, Fisher, A.J. | | Deposit date: | 2005-11-07 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytidine 5'-Monophosphate (CMP)-Induced Structural Changes in a Multifunctional Sialyltransferase from Pasteurella multocida
Biochemistry, 45, 2006
|
|
2EX0
 
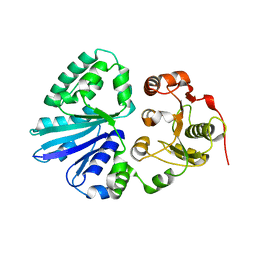 | | Crystal structure of multifunctional sialyltransferase from Pasteurella Multocida | | Descriptor: | a2,3-sialyltransferase, a2,6-sialyltransferase | | Authors: | Ni, L, Sun, M, Chen, X, Fisher, A.J. | | Deposit date: | 2005-11-07 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cytidine 5'-Monophosphate (CMP)-Induced Structural Changes in a Multifunctional Sialyltransferase from Pasteurella multocida
Biochemistry, 45, 2006
|
|
8IX3
 
 | | Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (local refinement) | | Descriptor: | BA.4/5 variant spike protein, heavy chain of 1G11, light chain of 1G11 | | Authors: | Sun, H, Jiang, Y, Zheng, Z, Zheng, Q, Li, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for broad neutralization of human antibody against Omicron sublineages and evasion by XBB variant.
J.Virol., 97, 2023
|
|
2IHK
 
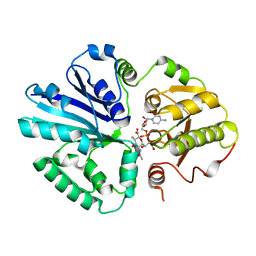 | |
2IHJ
 
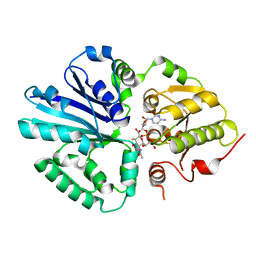 | |
2IIQ
 
 | |
2ILV
 
 | |
2IHZ
 
 | |
2II6
 
 | |
2IIB
 
 | |
4R83
 
 | | Crystal structure of Sialyltransferase from Photobacterium damsela | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Fisher, A.J, Chen, X, Li, Y, Huynh, N. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4R9V
 
 | | Crystal structure of sialyltransferase from photobacterium damselae, residues 113-497 corresponding to the gt-b domain | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Li, Y, Huynh, N, Chen, X, Fisher, A.J. | | Deposit date: | 2014-09-08 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4R84
 
 | | Crystal structure of Sialyltransferase from Photobacterium damsela with CMP-3F(a)Neu5Ac bound | | Descriptor: | CALCIUM ION, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, Sialyltransferase 0160 | | Authors: | Fisher, A.J, Chen, X, Li, Y, Huynh, N. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4RHR
 
 | | Crystal structure of PltB | | Descriptor: | ACETATE ION, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Wang, J, Galan, J. | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-29 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Host adaptation of a bacterial toxin from the human pathogen salmonella typhi.
Cell(Cambridge,Mass.), 159, 2014
|
|
4RHS
 
 | | Crystal structure of GD2 bound PltB | | Descriptor: | ACETATE ION, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Wang, J, Galan, J. | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9211 Å) | | Cite: | Host adaptation of a bacterial toxin from the human pathogen salmonella typhi.
Cell(Cambridge,Mass.), 159, 2014
|
|
7XRQ
 
 | |
