7FDV
 
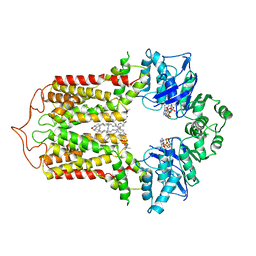 | | Cryo-EM structure of the human cholesterol transporter ABCG1 in complex with cholesterol | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 1, CHOLESTEROL, ... | | 著者 | Xu, D, Li, Y.Y, Yang, F.R, Sun, C.R, Pan, J.H, Wang, L, Chen, Z.P, Fang, S.C, Yao, X.B, Hou, W.T, Zhou, C.Z, Chen, Y. | | 登録日 | 2021-07-18 | | 公開日 | 2022-06-22 | | 最終更新日 | 2022-06-29 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Structure and transport mechanism of the human cholesterol transporter ABCG1.
Cell Rep, 38, 2022
|
|
7Y3I
 
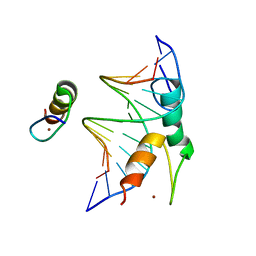 | | Structure of DNA bound SALL4 | | 分子名称: | DNA (12-mer), Sal-like protein 4, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-10 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-30 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3M
 
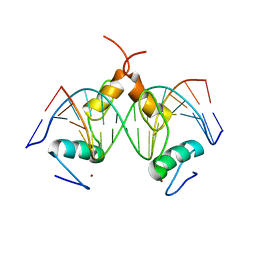 | | Structure of SALL4 ZFC1 bound with 16 bp AT-rich dsDNA | | 分子名称: | DNA (16-mer), Sal-like protein 4, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.723 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3K
 
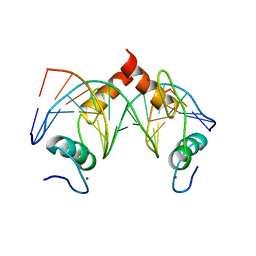 | | Structure of SALL4 ZFC4 bound with 16 bp AT-rich dsDNA | | 分子名称: | DNA (16-mer), Sal-like protein 4, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3L
 
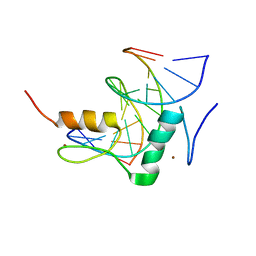 | | Structure of SALL3 ZFC4 bound with 12 bp AT-rich dsDNA | | 分子名称: | DNA (12-mer), Sal-like protein 3, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7XHO
 
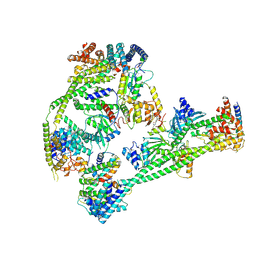 | | Structure of human inner kinetochore CCAN complex | | 分子名称: | CENP-W, Centromere protein C, Centromere protein H, ... | | 著者 | Tian, T, Wang, C.L, Yang, Z.S, Sun, L.F, Zang, J.Y. | | 登録日 | 2022-04-09 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-01-25 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
7XHN
 
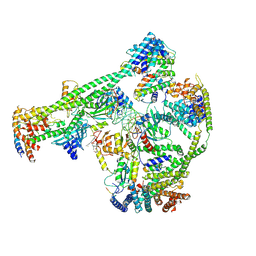 | | Structure of human inner kinetochore CCAN-DNA complex | | 分子名称: | CENP-W, Centromere protein C, Centromere protein H, ... | | 著者 | Sun, L.F, Tian, T, Wang, C.L, Yang, Z.S, Zang, J.Y. | | 登録日 | 2022-04-09 | | 公開日 | 2023-01-25 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
8JAR
 
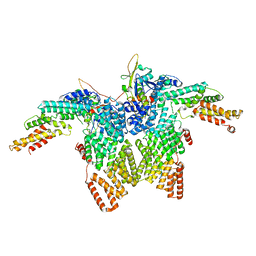 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (dimer) | | 分子名称: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | 著者 | Zhao, S, Zhang, K, Xu, C. | | 登録日 | 2023-05-07 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAV
 
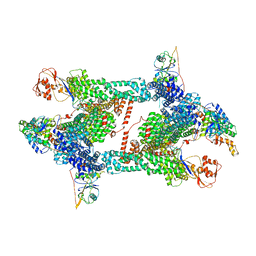 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (tetramer) | | 分子名称: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Zhao, S, Zhang, K, Xu, C. | | 登録日 | 2023-05-07 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAU
 
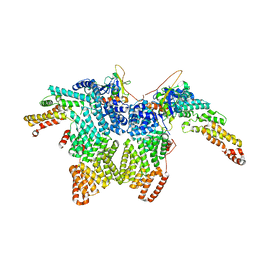 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (dimer) | | 分子名称: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | 著者 | Zhao, S, Zhang, K, Xu, C. | | 登録日 | 2023-05-07 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAQ
 
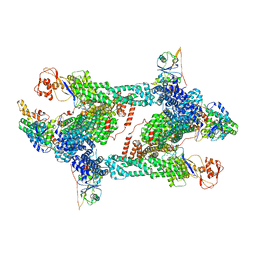 | | Structure of CRL2APPBP2 bound with RxxGP degron (tetramer) | | 分子名称: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Zhao, S, Zhang, K, Xu, C. | | 登録日 | 2023-05-06 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAS
 
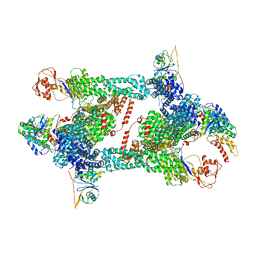 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (tetramer) | | 分子名称: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Zhao, S, Zhang, K, Xu, C. | | 登録日 | 2023-05-07 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAL
 
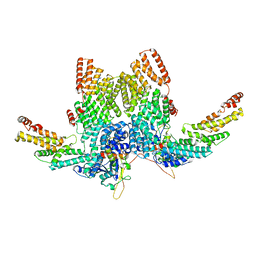 | | Structure of CRL2APPBP2 bound with RxxGP degron (dimer) | | 分子名称: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | 著者 | Zhao, S, Zhang, K, Xu, C. | | 登録日 | 2023-05-06 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6L0X
 
 | | The First Tudor Domain of PHF20L1 | | 分子名称: | CITRIC ACID, GLYCEROL, PHD finger protein 20-like protein 1 | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6H02
 
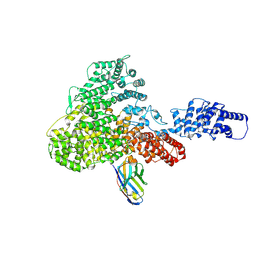 | |
6L1P
 
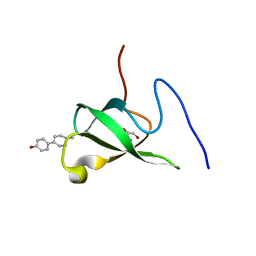 | | Crystal structure of PHF20L1 in complex with Hit 1 | | 分子名称: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-29 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.231 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1C
 
 | |
6L1I
 
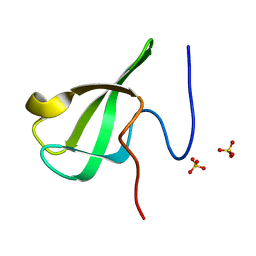 | |
6L10
 
 | | PHF20L1 Tudor1 - MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1F
 
 | |
7VKH
 
 | |
7VKG
 
 | |
7YF2
 
 | |
7Y9C
 
 | |
7YF4
 
 | |
