4L81
 
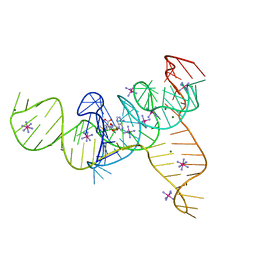 | | Structure of the SAM-I/IV riboswitch (env87(deltaU92, deltaG93)) | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Trausch, J.J, Reyes, F.E, Edwards, A.L, Batey, R.T. | | Deposit date: | 2013-06-15 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for diversity in the SAM clan of riboswitches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5U6V
 
 | | X-ray crystal structure of 1,2,3-triazolobenzodiazepine in complex with BRD2(D2) | | Descriptor: | 1,2-ETHANEDIOL, 5-[7-(4-chlorophenyl)-1-methyl-6,7-dihydro-5H-[1,2,3]triazolo[1,5-d][1,4]benzodiazepin-9-yl]pyridin-2-amine, Bromodomain-containing protein 2 | | Authors: | Hatfaludi, T, Sharp, P.P, Garnier, J.-M, Burns, C.J, Czabotar, P.E. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | Design, Synthesis, and Biological Activity of 1,2,3-Triazolobenzodiazepine BET Bromodomain Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5V84
 
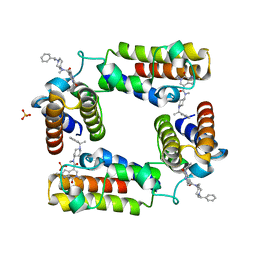 | | CECR2 in complex with Cpd6 (6-allyl-N,2-dimethyl-7-oxo-N-(1-(1-phenylethyl)piperidin-4-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4-carboxamide) | | Descriptor: | Cat eye syndrome critical region protein 2, N,2-dimethyl-7-oxo-N-{1-[(1S)-1-phenylethyl]piperidin-4-yl}-6-(prop-2-en-1-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4- carboxamide, SULFATE ION | | Authors: | Murray, J.M, Kiefer, J.R, Jayaran, H, Bellon, S, Boy, F. | | Deposit date: | 2017-03-21 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | GNE-886: A Potent and Selective Inhibitor of the Cat Eye Syndrome Chromosome Region Candidate 2 Bromodomain (CECR2).
ACS Med Chem Lett, 8, 2017
|
|
5W3M
 
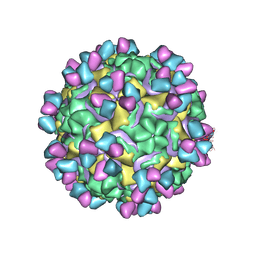 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:1, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3O
 
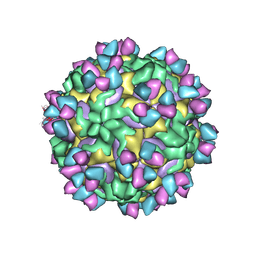 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, empty particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3L
 
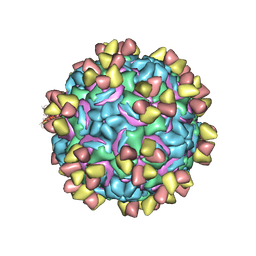 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (4 degrees Celsius, molar ratio 1:3, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3E
 
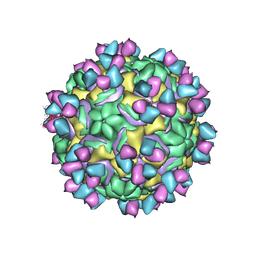 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
4OQU
 
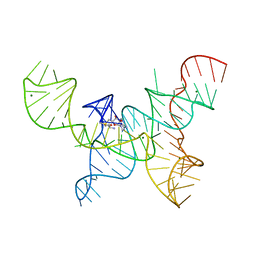 | | Structure of the SAM-I/IV riboswitch (env87(deltaU92)) | | Descriptor: | MAGNESIUM ION, S-ADENOSYLMETHIONINE, SAM-I/IV riboswitch | | Authors: | Trausch, J.J, Reyes, F.E, Edwards, A.L, Batey, R.T. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for diversity in the SAM clan of riboswitches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6BVG
 
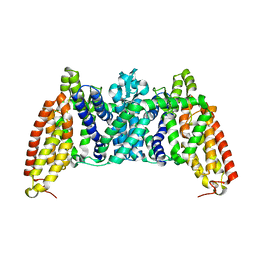 | | Crystal structure of bcMalT T280C-E54C crosslinked by divalent mercury | | Descriptor: | MERCURY (II) ION, Protein-N(Pi)-phosphohistidine-sugar phosphotransferase (Enzyme II of the phosphotransferase system) (PTS system glucose-specific IIBC component), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ren, Z, Zhou, M. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an EIIC sugar transporter trapped in an inward-facing conformation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3O39
 
 | |
6AY2
 
 | | Structure of CathB with covalently linked Compound 28 | | Descriptor: | Cathepsin B, N~1~-[(2S)-1-amino-5-(carbamoylamino)pentan-2-yl]-N'~1~-[(1R)-1-(thiophen-3-yl)ethyl]cyclobutane-1,1-dicarboxamide | | Authors: | Kiefer, J.R, Steinbacher, S. | | Deposit date: | 2017-09-07 | | Release date: | 2017-12-27 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Peptidomimetic Antibody-Drug Conjugate Linkers with Enhanced Protease Specificity.
J. Med. Chem., 61, 2018
|
|
7WY8
 
 | | ADGRL3/Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling
Mol.Cell, 82, 2022
|
|
7WYB
 
 | | ADGRL3/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and G q , G s , G i , and G 12 coupling.
Mol.Cell, 82, 2022
|
|
7WY5
 
 | | ADGRL3/Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
7X10
 
 | | ADGRL3/miniG12 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
7CKH
 
 | | Crystal structure of TMSiPheRS | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79492676 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
7CKG
 
 | | Crystal structure of TMSiPheRS complexed with TMSiPhe | | Descriptor: | 4-(trimethylsilyl)-L-phenylalanine, Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
7Y50
 
 | | Class I diterpene synthase (CyS) from Streptomyces cattleya | | Descriptor: | Putative glutamate dehydrogenase/leucine dehydrogenase | | Authors: | Xing, B, Ma, M. | | Deposit date: | 2022-06-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Based Mutagenesis of Cattleyene Synthase Leads to the Generation of Rearranged Polycyclic Diterpenes.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y87
 
 | |
7Y88
 
 | | class I diterpene synthase (CyS-GGPP-Mg2+) from Streptomyces cattleya | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, MAGNESIUM ION, Putative glutamate dehydrogenase/leucine dehydrogenase | | Authors: | Xing, B, Ma, M. | | Deposit date: | 2022-06-23 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure Based Mutagenesis of Cattleyene Synthase Leads to the Generation of Rearranged Polycyclic Diterpenes.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4H42
 
 | | Synthesis of a Weak Basic uPA Inhibitor and Crystal Structure of Complex with uPA | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, N-[(2-amino-1,3-benzothiazol-6-yl)carbonyl]glycine, Urokinase-type plasminogen activator | | Authors: | Yu, H.-Y, Gao, D, Zhang, X, Jiang, L.-G, Hong, Z.-B, Yuan, C, Fang, X, Wang, J.-D, Huang, M.-D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Synthesis of a Weak Basic uPA Inhibitor and Crystal Structure of Complex with uPA
CHIN.J.STRUCT.CHEM., 32, 2013
|
|
7F35
 
 | | Crystal structure of anti S-gatifloxacin antibody Fab fragment in complex with S-gatifloxacin | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-6-fluoro-8-methoxy-7-[(3S)-3-methylpiperazin-1-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, Antibody Fab fragment heavy chain, ... | | Authors: | Wang, L.T, Jiao, W.Y, Shen, X, Lei, H.T. | | Deposit date: | 2021-06-15 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational adaptability determining antibody recognition to distomer: structure analysis of enantioselective antibody against chiral drug gatifloxacin
Rsc Adv, 11, 2021
|
|
7F2S
 
 | | Crystal structure of anti S-gatifloxacin antibody Fab fragment apo form | | Descriptor: | 1,2-ETHANEDIOL, Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, ... | | Authors: | Wang, L.T, Jiao, W.Y, Shen, X, Lei, H.T. | | Deposit date: | 2021-06-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Conformational adaptability determining antibody recognition to distomer: structure analysis of enantioselective antibody against chiral drug gatifloxacin
Rsc Adv, 11, 2021
|
|
7F0A
 
 | |
