4ZHM
 
 | |
4ZHL
 
 | |
6ZGI
 
 | | Furin Cleaved Spike Protein of SARS-CoV-2 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZGF
 
 | | Spike Protein of RaTG13 Bat Coronavirus in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZGH
 
 | |
6ZGE
 
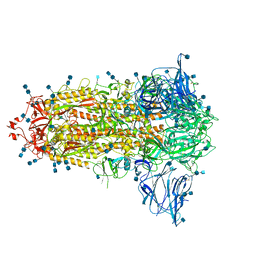 | | Uncleavable Spike Protein of SARS-CoV-2 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZGG
 
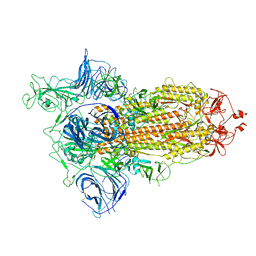 | |
7A98
 
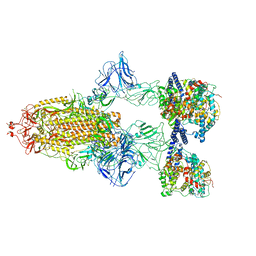 | | SARS-CoV-2 Spike Glycoprotein with 3 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A92
 
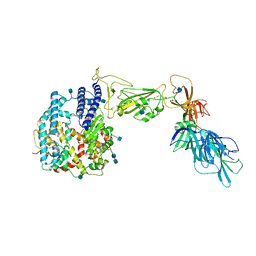 | | Dissociated S1 domain of SARS-CoV-2 Spike bound to ACE2 (Unmasked Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-30 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A94
 
 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A95
 
 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Clockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A91
 
 | | Dissociated S1 domain of SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A96
 
 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Anticlockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A97
 
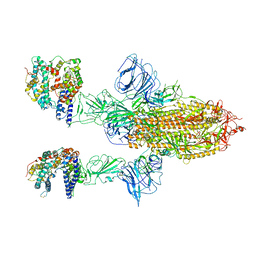 | | SARS-CoV-2 Spike Glycoprotein with 2 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A93
 
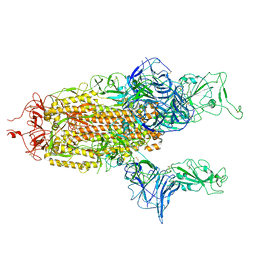 | |
7BNN
 
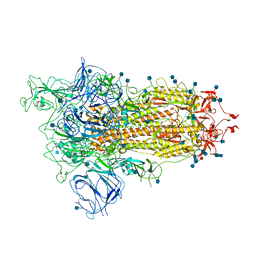 | | Open conformation of D614G SARS-CoV-2 spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BBH
 
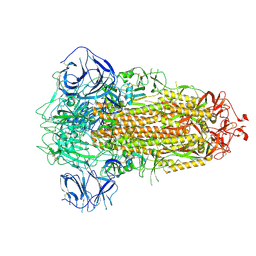 | | Structure of Coronavirus Spike from Smuggled Guangdong Pangolin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface glycoprotein | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7BNO
 
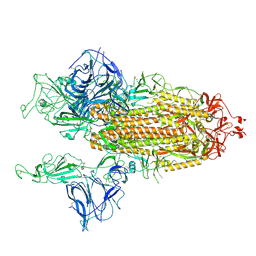 | | Open conformation of D614G SARS-CoV-2 spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BNM
 
 | | Closed conformation of D614G SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6A8O
 
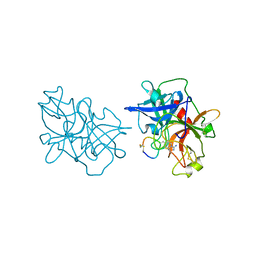 | | Crystal structures of the serine protease domain of murine plasma kallikrein with peptide inhibitor mupain-1-16 | | Descriptor: | Plasma kallikrein, alpha-D-mannopyranose, peptide inhibitor,, ... | | Authors: | Xu, M, Jiang, L, Huang, M. | | Deposit date: | 2018-07-09 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of plasma kallikrein reveals the unusual flexibility of the S1 pocket triggered by Glu217.
Febs Lett., 592, 2018
|
|
5ZAT
 
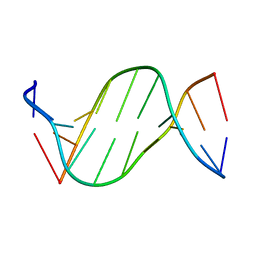 | | Crystal structure of 5-carboxylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(CAC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
7W53
 
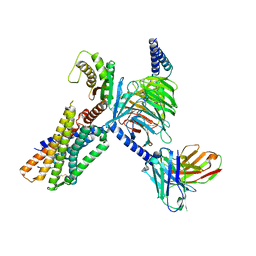 | | Cryo-EM structure of the neuromedin U-bound neuromedin U receptor 1-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | You, C, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-20 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the peptide selectivity and activation of human neuromedin U receptors.
Nat Commun, 13, 2022
|
|
7W57
 
 | | Cryo-EM structure of the neuromedin S-bound neuromedin U receptor 2-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | You, C, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-20 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the peptide selectivity and activation of human neuromedin U receptors.
Nat Commun, 13, 2022
|
|
7W55
 
 | | Cryo-EM structure of the neuromedin U-bound neuromedin U receptor 2-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | You, C, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-20 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the peptide selectivity and activation of human neuromedin U receptors.
Nat Commun, 13, 2022
|
|
7W56
 
 | | Cryo-EM structure of the neuromedin S-bound neuromedin U receptor 1-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | You, C, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-20 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the peptide selectivity and activation of human neuromedin U receptors.
Nat Commun, 13, 2022
|
|
