2X5G
 
 | | Crystal structure of the ORF131L51M mutant from Sulfolobus islandicus rudivirus 1 | | Descriptor: | CHLORIDE ION, MALONATE ION, ORF 131 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Naismith, J.H, White, M.F. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X3G
 
 | | Crystal Structure of the hypothetical protein ORF119 from Sulfolobus islandicus rod-shaped virus 1 | | Descriptor: | SIRV1 HYPOTHETICAL PROTEIN ORF119 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X3O
 
 | | Crystal Structure of the Hypothetical Protein PA0856 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, GLYCEROL, HYPOTHETICAL PROTEIN PA0856 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3M
 
 | | Crystal Structure of Hypothetical Protein ORF239 from Pyrobaculum Spherical Virus | | Descriptor: | HYPOTHETICAL PROTEIN ORF239 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4L
 
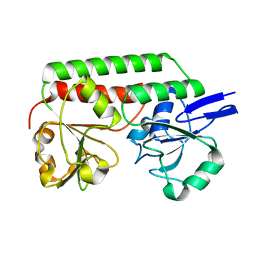 | | Crystal structure of DesE, a ferric-siderophore receptor protein from Streptomyces coelicolor | | Descriptor: | FERRIC-SIDEROPHORE RECEPTOR PROTEIN | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X7I
 
 | | Crystal structure of mevalonate kinase from methicillin-resistant Staphylococcus aureus MRSA252 | | Descriptor: | CHLORIDE ION, CITRIC ACID, MEVALONATE KINASE | | Authors: | Oke, M, Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-27 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3D
 
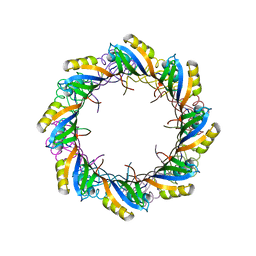 | | Crystal Structure of SSo6206 from Sulfolobus solfataricus P2 | | Descriptor: | SSO6206 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, McMahon, S.A, McEwan, A.R, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: targets, methods and outputs.
J. Struct. Funct. Genomics, 11, 2010
|
|
2X5H
 
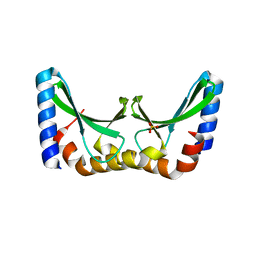 | | Crystal structure of the ORF131 L26M L51M double mutant from Sulfolobus islandicus rudivirus 1 | | Descriptor: | ORF 131, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Naismith, J.H, White, M.F. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X5D
 
 | | Crystal Structure of a probable aminotransferase from Pseudomonas aeruginosa | | Descriptor: | PROBABLE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
3ZFV
 
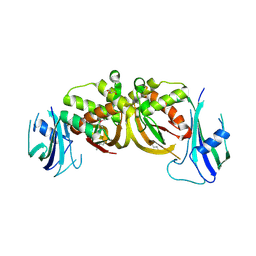 | | Crystal structure of an archaeal CRISPR-associated Cas6 nuclease | | Descriptor: | CRISPR-ASSOCIATED ENDORIBONUCLEASE CAS6 1, GLYCEROL | | Authors: | Reeks, J, Liu, H, White, M.F, Naismith, J.H. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-03 | | Last modified: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Dimeric Crenarchaeal Cas6 Enzyme with an Atypical Active Site for Crispr RNA Processing
Biochem.J., 452, 2013
|
|
3ZC4
 
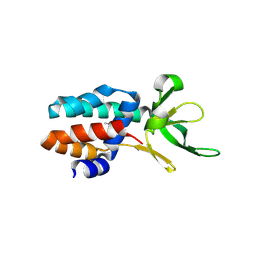 | | The structure of Csa5 from Sulfolobus solfataricus. | | Descriptor: | DI(HYDROXYETHYL)ETHER, SSO1398 | | Authors: | Reeks, J, Anderson, L, White, M.F, Naismith, J.H. | | Deposit date: | 2012-11-15 | | Release date: | 2013-02-20 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of the Archaeal Cascade Subunit Csa5: Relating the Small Subunits of Crispr Effector Complexes.
RNA Biol., 10, 2013
|
|
1OB9
 
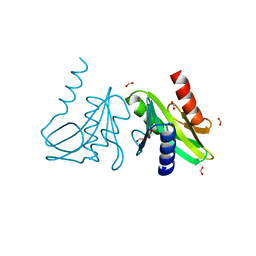 | | Holliday Junction Resolving Enzyme | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, HOLLIDAY JUNCTION RESOLVASE | | Authors: | Middleton, C.L, Parker, J.L, Richard, D.J, White, M.F, Bond, C.S. | | Deposit date: | 2003-01-28 | | Release date: | 2004-10-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Recognition and Catalysis by the Holliday Junction Resolving Enzyme Hje.
Nucleic Acids Res., 32, 2004
|
|
1OB8
 
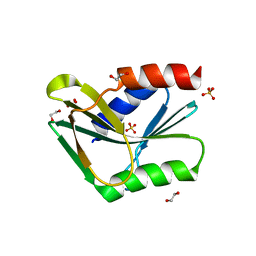 | | Holliday Junction Resolving Enzyme | | Descriptor: | 1,2-ETHANEDIOL, HOLLIDAY-JUNCTION RESOLVASE, SULFATE ION | | Authors: | Middleton, C.L, Parker, J.L, Richard, D.J, White, M.F, Bond, C.S. | | Deposit date: | 2003-01-28 | | Release date: | 2004-10-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Recognition and Catalysis by the Holliday Junction Resolving Enzyme Hje.
Nucleic Acids Res., 32, 2004
|
|
2XVO
 
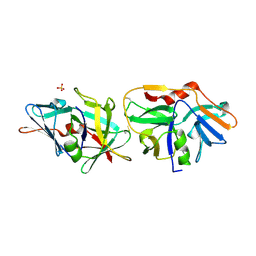 | | SSO1725, a protein involved in the CRISPR/Cas pathway | | Descriptor: | BETA-MERCAPTOETHANOL, SSO1725, SULFATE ION | | Authors: | Reeks, J, Liu, H, Naismith, J, White, M, McMahon, S. | | Deposit date: | 2010-10-26 | | Release date: | 2010-12-29 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure and Mechanism of the Cmr Complex for Crispr-Mediated Antiviral Immunity.
Mol.Cell, 45, 2012
|
|
7QDA
 
 | | Crystal structure of CalpL | | Descriptor: | CalpL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-16 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
8B0R
 
 | | Structure of the CalpL/cA4 complex | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), SMODS-associated and fused to various effectors domain-containing protein, SULFATE ION, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
8B0U
 
 | | Structure of the CalpL/T10 complex | | Descriptor: | CalpT10, GLYCEROL, SAVED domain-containing protein, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
5H8C
 
 | | Truncated XPD | | Descriptor: | IRON/SULFUR CLUSTER, XPD/Rad3 related DNA helicase | | Authors: | Naismith, J.H, Constantinescu, D. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Mechanism of DNA loading by the DNA repair helicase XPD.
Nucleic Acids Res., 44, 2016
|
|
5H8W
 
 | | XPD mechanism | | Descriptor: | ATP-dependent DNA helicase Ta0057, DNA (5'-D(P*TP*AP*CP*GP*A)-3'), IRON/SULFUR CLUSTER, ... | | Authors: | Naismith, J.H, Constantinescu, D. | | Deposit date: | 2015-12-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of DNA loading by the DNA repair helicase XPD.
Nucleic Acids Res., 44, 2016
|
|
4TKD
 
 | | Sulfolobus solfataricus HJC mutants | | Descriptor: | Holliday junction resolvase Hjc | | Authors: | Bond, C.S. | | Deposit date: | 2014-05-26 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Unengineering: Reducing the Crystallisability of Sulfolobus solfataricus Hjc
Aust.J.Chem., 67, 2014
|
|
4TKK
 
 | | Sulfolobus solfataricus HJC mutants | | Descriptor: | Holliday junction resolvase Hjc | | Authors: | Bond, C.S. | | Deposit date: | 2014-05-27 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Unengineering: Reducing the Crystallisability of Sulfolobus solfataricus Hjc
Aust.J.Chem., 67, 2014
|
|
3PS0
 
 | | The structure of the CRISPR-associated protein, csa2, from Sulfolobus solfataricus | | Descriptor: | CRISPR-Associated protein, CSA2 | | Authors: | Lintner, N.G, Sdano, M, Young, M.J, Lawrence, C.M. | | Deposit date: | 2010-11-30 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE).
J.Biol.Chem., 286, 2011
|
|
3BU5
 
 | |
3BU6
 
 | |
2JG5
 
 | | CRYSTAL STRUCTURE OF A PUTATIVE PHOSPHOFRUCTOKINASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | FRUCTOSE 1-PHOSPHATE KINASE | | Authors: | Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Dorward, M, McMahon, S.A, Oke, M, Powers, H, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
