7R8L
 
 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody C099 and CR3022 | | Descriptor: | C099 Fab Heavy Chain, C099 Fab Light Chain, CR3022 Fab heavy chain, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8N
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C051 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C051 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8M
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C032 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLaitsch, A.T, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7MY2
 
 | |
7MY3
 
 | |
8TV0
 
 | | XptA2 wild type | | Descriptor: | XptA2 | | Authors: | Martin, C.L, Aller, S.G. | | Deposit date: | 2023-08-17 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Insecticidal Toxin Complex Subunit XptA2 Highlight Roles for Flexible Domains.
Int J Mol Sci, 24, 2023
|
|
8TQE
 
 | | XptA2 wild type | | Descriptor: | XptA2 | | Authors: | Martin, C.L, Binshtein, E.M, Aller, S.G. | | Deposit date: | 2023-08-07 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the Insecticidal Toxin Complex Subunit XptA2 Highlight Roles for Flexible Domains.
Int J Mol Sci, 24, 2023
|
|
7CU5
 
 | | N-Glycosylation of PD-1 and glycosylation dependent binding of PD-1 specific monoclonal antibody camrelizumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Programmed cell death protein 1, ... | | Authors: | Liu, K.F, Tan, S.G, Jin, W.J, Guan, J.W, Wang, W.L, Sun, H, Qi, J.X, Yan, J.H, Chai, Y, Wang, Z.F, Chu, X.D, Gao, G.F. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | N-glycosylation of PD-1 promotes binding of camrelizumab.
Embo Rep., 21, 2020
|
|
8W6P
 
 | | Crystal structure of dimeric murine SMPDL3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Zhang, C, Liu, P, Fan, S, Hou, Y. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
8W6R
 
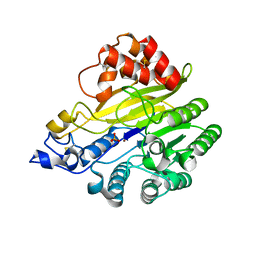 | | murine SMPDL3A bound to sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Zhang, C, Liu, P, Fan, S. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
7DVB
 
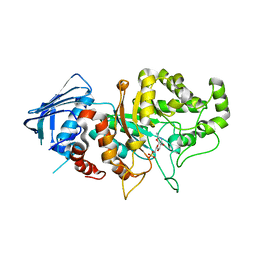 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DUP
 
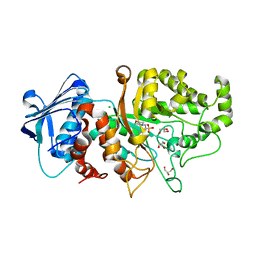 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVA
 
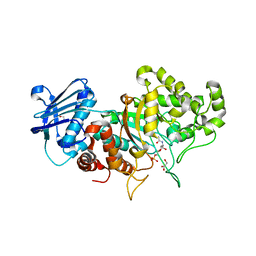 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
4XX1
 
 | | Low resolution structure of LCAT in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 heavy chain, Fab1 light chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
4XWG
 
 | | Crystal Structure of LCAT (C31Y) in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 Heavy Chain, Fab1 Light Chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-28 | | Release date: | 2015-07-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
4YNE
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 6-[(2R)-2-(3-fluorophenyl)pyrrolidin-1-yl]-3-(pyridin-2-yl)imidazo[1,2-b]pyridazine, GLYCEROL, High affinity nerve growth factor receptor, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0229 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4YMJ
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-[6-(benzylamino)imidazo[1,2-b]pyridazin-3-yl]benzonitrile, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
8TBN
 
 | | Tricomplex of RMC-7977, KRAS G12S, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 2024
|
|
8TBH
 
 | | Tricomplex of RMC-7977, KRAS G12R, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Bar Ziv, T, Zhang, D, Tomlinson, A.C.A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 2024
|
|
8TBK
 
 | | Tricomplex of RMC-7977, KRAS G12C, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 2024
|
|
7BY7
 
 | | Bacteriophage SPO1 protein Gp46 | | Descriptor: | Putative gene 46 protein | | Authors: | Liu, B, Zhang, P. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Bacteriophage protein Gp46 is a cross-species inhibitor of nucleoid-associated HU proteins
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8TBL
 
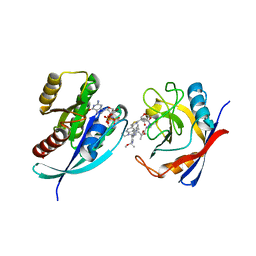 | | Tricomplex of RMC-7977, KRAS G12D, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 2024
|
|
8TBM
 
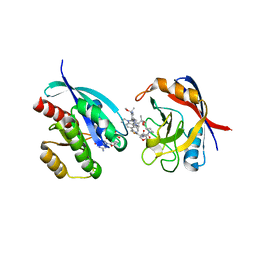 | | Tricomplex of RMC-7977, KRAS G12V, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GLYCEROL, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 2024
|
|
8TBJ
 
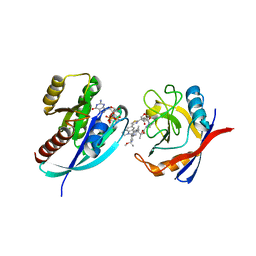 | | Tricomplex of RMC-7977, KRAS G12A, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 2024
|
|
8TBI
 
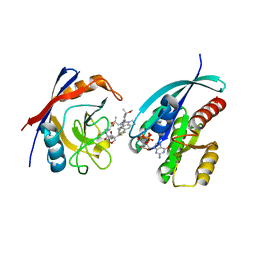 | | Tricomplex of RMC-7977, NRAS WT, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GTPase NRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 2024
|
|
