2P2O
 
 | | Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form | | Descriptor: | Maltose transacetylase | | Authors: | Liu, Z.J, Li, Y, Chen, L, Zhu, J, Rose, J.P, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Maltose Transacetylase from Geobacillus Kaustophilus at 1.8 Angstrom Resolution
To be Published
|
|
2PH3
 
 | | Crystal structure of 3-oxoacyl-[acyl carrier protein] reductase TTHA0415 from Thermus thermophilus | | Descriptor: | 3-oxoacyl-[acyl carrier protein] reductase | | Authors: | Swindell II, J.T, Chen, L, Zhu, J, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Chrzas, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of 3-oxoacyl-[acyl carrier protein] reductase TTHA0415 from Thermus thermophilus
To be Published
|
|
2QUF
 
 | | Crystal Structure of Transcription Factor AXXA-PF0095 from Pyrococcus furiosus | | Descriptor: | GLYCEROL, Transcription Factor PF0095 | | Authors: | Yang, H, Lipscomb, G.L, Scott, R.A, Rose, J.P, Wang, B.C. | | Deposit date: | 2007-08-05 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Transcription Factor AXXA-PF0095 from Pyrococcus furiosus
To be Published
|
|
2QLZ
 
 | |
2QVO
 
 | | Crystal structure of AF1382 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_1382 | | Authors: | Zhu, J, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Hao, X, Chen, L, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-08-08 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of AF1382 from Archaeoglobus fulgidus.
To be Published
|
|
4MN0
 
 | | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca2+-loaded apoprotein conformation state | | Descriptor: | Berovin, CALCIUM ION, MAGNESIUM ION | | Authors: | Liu, Z.J, Stepanyuk, G.A, Vysotski, E.S, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2013-09-09 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca(2+)-loaded apoprotein conformation state.
Biochim.Biophys.Acta, 1834, 2013
|
|
4PKM
 
 | | Crystal Structure of Bacillus thuringiensis Cry51Aa1 Protoxin at 1.65 Angstroms Resolution | | Descriptor: | Cry51Aa1, GLYCEROL, GLYCINE, ... | | Authors: | Xu, C, Chinte, U, Chen, L, Yao, Q, Zhou, D, Meng, Y, Li, L, Rose, J, Bi, L.J, Yu, Z, Sun, M, Wang, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Cry51Aa1: A potential novel insecticidal aerolysin-type beta-pore-forming toxin from Bacillus thuringiensis.
Biochem.Biophys.Res.Commun., 462, 2015
|
|
2HPW
 
 | | Green fluorescent protein from Clytia gregaria | | Descriptor: | Green fluorescent protein | | Authors: | Stepanyuk, G, Liu, Z.J, Vysotski, S.E, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Green Fluorescent Protein from Clytia Gregaria at 1.55 A resolution
To be Published
|
|
2HQ4
 
 | | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH1570 | | Authors: | Li, Y, Marshall, M, Chang, J, Zhao, M, Zhang, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii
To be Published
|
|
2HQ8
 
 | | Crystal structure of coelenterazine-binding protein from renilla muelleri in the ca loaded apo form | | Descriptor: | CALCIUM ION, Coelenterazine-binding protein ca-bound apo form | | Authors: | Stepanyuk, G, Liu, Z.J, Vysotski, E.S, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of coelenterazine-binding protein from Renilla muelleri at 1.7 A: why it is not a calcium-regulated photoprotein.
PHOTOCHEM.PHOTOBIOL.SCI., 7, 2008
|
|
2HQE
 
 | | Crystal structure of human P100 Tudor domain: Large fragment | | Descriptor: | P100 Co-activator tudor domain | | Authors: | Shah, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Liu, Z.J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a large fragment of the Human P100 Tudor Domain
To be Published
|
|
2HQX
 
 | | Crystal structure of human P100 tudor domain conserved region | | Descriptor: | P100 CO-ACTIVATOR TUDOR DOMAIN | | Authors: | Zhao, M, Liu, Z.J, Xu, H, Yang, J, Silvennoinen, O, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-19 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Human P100 Tudor Domain Conserved Region
To be Published
|
|
2HPS
 
 | | Crystal structure of coelenterazine-binding protein from Renilla Muelleri | | Descriptor: | C2-HYDROXY-COELENTERAZINE, GLYCEROL, coelenterazine-binding protein with bound coelenterazine | | Authors: | Stepanyuk, G, Liu, Z.J, Vysotski, E.S, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-17 | | Release date: | 2007-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of coelenterazine-binding protein from Renilla muelleri at 1.7 A: why it is not a calcium-regulated photoprotein.
PHOTOCHEM.PHOTOBIOL.SCI., 7, 2008
|
|
2HR5
 
 | | PF1283- Rubrerythrin from Pyrococcus furiosus iron bound form | | Descriptor: | FE (III) ION, Rubrerythrin | | Authors: | Dillard, B.D, Ruble, J.R, Chen, L, Liu, Z.J, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-19 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of iron bound Rubrerythrin from Pyrococcus Furiosus
To be Published
|
|
2HPV
 
 | | Crystal structure of FMN-Dependent azoreductase from Enterococcus faecalis | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Liu, Z.J, Chen, L, Chen, H, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-12 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Fmn-Dependent Azoreductase from Enterococcus faecalis at 2.00 A resolution
To be Published
|
|
2HQ1
 
 | | Crystal Structure of ORF 1438 a putative Glucose/ribitol dehydrogenase from Clostridium thermocellum | | Descriptor: | Glucose/ribitol dehydrogenase | | Authors: | Southeast Collaboratory for Structural Genomics (SECSG), Li, Y, Shaw, N, Xu, H, Cheng, C, Chen, L, Liu, Z.J, Rose, J.P, Wang, B.C. | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of ORF 1438 a putative Glucose/ ribitol dehydrogenase from Clostridium thermocellum
To be Published
|
|
2IA0
 
 | | Transcriptional Regulatory Protein PF0864 From Pyrococcus Furiosus a Member of the ASNC Family (PF0864) | | Descriptor: | GOLD ION, Putative HTH-type transcriptional regulator PF0864 | | Authors: | Yang, H, Chang, J, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal Structure of the Transcriptional Regulatory Protein PF0864: an Asnc Family member from Pyrococcus Furiosus
To be Published
|
|
2IEL
 
 | | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus | | Descriptor: | Hypothetical Protein TT0030 | | Authors: | Zhu, J, Huang, J, Stepanyuk, G, Chen, L, Chang, J, Zhao, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus AT 1.6 ANGSTROMS RESOLUTION
To be Published
|
|
2IC7
 
 | | Crystal Structure of Maltose Transacetylase from Geobacillus kaustophilus | | Descriptor: | Maltose transacetylase | | Authors: | Liu, Z.J, Li, Y, Chen, L, Zhu, J, Rose, J.P, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-12 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Maltose Transacetylase From Geobacillus kaustophilus at 1.78 Angstrom Resolution
To be Published
|
|
2ICU
 
 | | Crystal Structure of Hypothetical Protein YedK From Escherichia coli | | Descriptor: | Hypothetical protein yedK | | Authors: | Chen, L, Liu, Z.J, Li, Y, Zhao, M, Rose, J, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-13 | | Release date: | 2006-11-07 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Hypothetical Protein YedK From Escherichia coli
To be Published
|
|
2IDG
 
 | | Crystal Structure of hypothetical protein AF0160 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0160 | | Authors: | Zhao, M, Zhang, M, Chang, J, Chen, L, Xu, H, Li, Y, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-11-14 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Hypothetical Protein AF0160 from Archaeoglobus fulgidus at 2.69 Angstrom resolution
To be Published
|
|
1GGG
 
 | |
1JK6
 
 | | UNCOMPLEXED DES 1-6 BOVINE NEUROPHYSIN | | Descriptor: | NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2003
|
|
1JK4
 
 | | DES 1-6 BOVINE NEUROPHYSIN II COMPLEX WITH VASOPRESSIN | | Descriptor: | CADMIUM ION, Lys Vasopressin, NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2001
|
|
1WDN
 
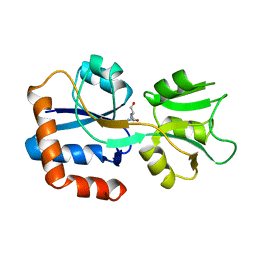 | | GLUTAMINE-BINDING PROTEIN | | Descriptor: | GLUTAMINE, GLUTAMINE BINDING PROTEIN | | Authors: | Sun, Y.-J, Rose, J, Wang, B.-C, Hsiao, C.-D. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of glutamine-binding protein complexed with glutamine at 1.94 A resolution: comparisons with other amino acid binding proteins.
J.Mol.Biol., 278, 1998
|
|
