7KWZ
 
 | | TDP-43 LCD amyloid fibrils | | Descriptor: | Isoform 2 of TAR DNA-binding protein 43 | | Authors: | Li, Q, Babinchak, W.M, Surewicz, W.K. | | Deposit date: | 2020-12-02 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43.
Nat Commun, 12, 2021
|
|
5ZL4
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 wihout its lid in complex with GF2 | | Descriptor: | DFA-IIIase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZL5
 
 | | Crystal structure of DFA-IIIase mutant C387A from Arthrobacter chlorophenolicus A6 | | Descriptor: | DFA-IIIase C387A mutant, GLYCEROL | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKW
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with GF2 | | Descriptor: | DFA-IIIase, alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZLA
 
 | | Crystal structure of mutant C387A of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-III | | Descriptor: | (2R,3'S,4'S,4aR,5'R,6R,7R,7aS)-4a,5',6-tris(hydroxymethyl)spiro[3,6,7,7a-tetrahydrofuro[2,3-b][1,4]dioxine-2,2'-oxolane ]-3',4',7-triol, DFA-IIIase C387A mutant | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-27 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKU
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-III | | Descriptor: | (2R,3'S,4'S,4aR,5'R,6R,7R,7aS)-4a,5',6-tris(hydroxymethyl)spiro[3,6,7,7a-tetrahydrofuro[2,3-b][1,4]dioxine-2,2'-oxolane ]-3',4',7-triol, DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKY
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 without its lid | | Descriptor: | DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKS
 
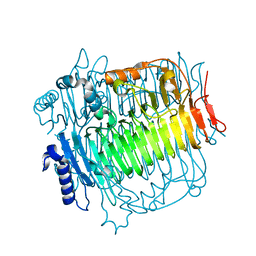 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 | | Descriptor: | DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
6BI6
 
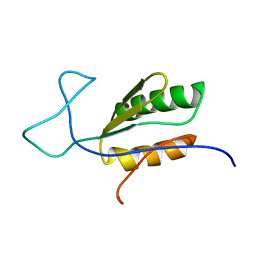 | | Solution NMR structure of uncharacterized protein YejG | | Descriptor: | Uncharacterized protein YejG | | Authors: | Mohanty, B, Finn, T.J, Macindoe, I, Zhong, J, Patrick, W.M, Mackay, J.P. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The uncharacterized bacterial protein YejG has the same architecture as domain III of elongation factor G.
Proteins, 87, 2019
|
|
2FTL
 
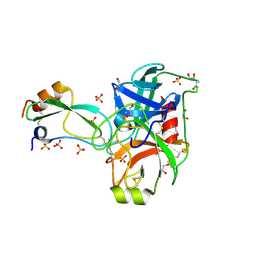 | | Crystal structure of trypsin complexed with BPTI at 100K | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Hanson, W.M, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rigidification of a Flexible Protease Inhibitor Variant upon Binding to Trypsin.
J.Mol.Biol., 366, 2007
|
|
2FV7
 
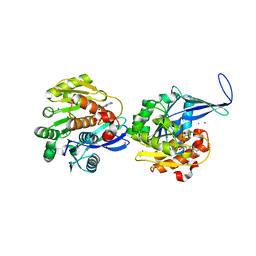 | | Crystal structure of human ribokinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribokinase, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-30 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human ribokinase
to be published
|
|
2FTM
 
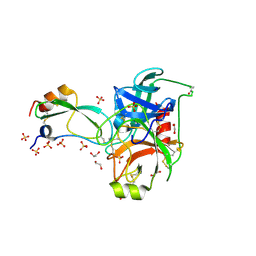 | | Crystal structure of trypsin complexed with the BPTI variant (Tyr35->Gly) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Hanson, W.M, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rigidification of a Flexible Protease Inhibitor Variant upon Binding to Trypsin.
J.Mol.Biol., 366, 2007
|
|
2H16
 
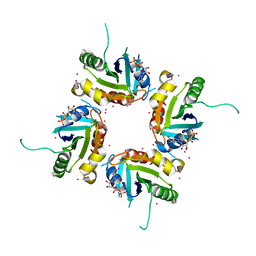 | | Structure of human ADP-ribosylation factor-like 5 (ARL5) | | Descriptor: | ADP-ribosylation factor-like protein 5A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 5 (ARL5)
To be Published
|
|
2H17
 
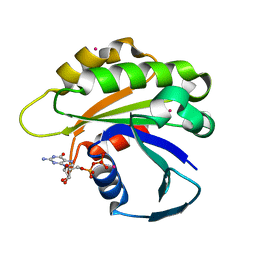 | | Structure of human ADP-ribosylation factor-like 5 (ARL5) | | Descriptor: | ADP-ribosylation factor-like protein 5A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 5 (ARL5)
To be Published
|
|
2HIN
 
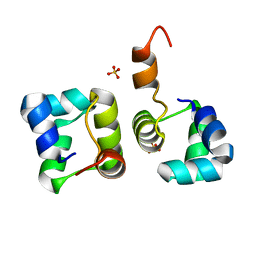 | | Structure of N15 Cro at 1.05 A: an ortholog of lambda Cro with a completely different but equally effective dimerization mechanism | | Descriptor: | Repressor protein, SULFATE ION | | Authors: | Dubrava, M.S, Ingram, W.M, Roberts, S.A, Weichsel, A, Montfort, W.R, Cordes, M.H. | | Deposit date: | 2006-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | N15 Cro and lambda Cro: orthologous DNA-binding domains with completely different but equally effective homodimer interfaces.
Protein Sci., 17, 2008
|
|
2HLZ
 
 | | Crystal Structure of human ketohexokinase | | Descriptor: | Ketohexokinase, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-10 | | Release date: | 2006-08-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of human ketohexokinase (CASP Target)
To be Published
|
|
2I7Q
 
 | | Crystal structure of Human Choline Kinase A | | Descriptor: | CHLORIDE ION, Choline kinase alpha, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Wasney, G, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Human Choline Kinase A
To be Published
|
|
2IG7
 
 | | Crystal structure of Human Choline Kinase B | | Descriptor: | Choline/ethanolamine kinase, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Wasney, G, Landry, R, Vedadi, M, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Human Choline Kinase B
To be Published
|
|
2JIH
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (complex-form) | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, ADAMTS-1, CADMIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains.
J. Mol. Biol., 373, 2007
|
|
2JC3
 
 | | Structure of O-Acetylserine Sulfhydrylase B from Salmonella Typhimurium | | Descriptor: | O-ACETYLSERINE SULFHYDRYLASE B, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chattopadhyay, A, Rabeh, W.M, Speroni, F, Meier, M, Ivaninskii, S, Mozzarelli, A, Burkhard, P, Cook, P.F. | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, Mechanism, and Conformational Dynamics of O-Acetylserine Sulfhydrylase from Salmonella Typhimurium: Comparison of a and B Isozymes.
Biochemistry, 46, 2007
|
|
2L9Q
 
 | |
2L15
 
 | | Solution Structure of Cold Shock Protein CspA Using Combined NMR and CS-Rosetta method | | Descriptor: | Cold shock protein CspA | | Authors: | Tang, Y, Schneider, W.M, Shen, Y, Raman, S, Inouye, M, Baker, D, Roth, M.J, Montelione, G.T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Fully automated high-quality NMR structure determination of small (2)H-enriched proteins.
J Struct Funct Genomics, 11, 2010
|
|
3I6A
 
 | | Human GST A1-1 GIMF mutant with Glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Balogh, L.M, Le Trong, I, Stenkamp, R.E, Atkins, W.M. | | Deposit date: | 2009-07-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural analysis of a glutathione transferase A1-1 mutant tailored for high catalytic efficiency with toxic alkenals.
Biochemistry, 48, 2009
|
|
3I69
 
 | | Apo Glutathione Transferase A1-1 GIMF-helix mutant | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Balogh, L.M, Le Trong, I, Stenkamp, R.E, Atkins, W.M. | | Deposit date: | 2009-07-06 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural analysis of a glutathione transferase A1-1 mutant tailored for high catalytic efficiency with toxic alkenals.
Biochemistry, 48, 2009
|
|
3IK5
 
 | |
