7SZ9
 
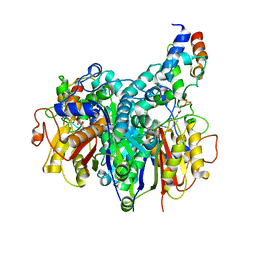 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C16:1-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N~3~-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-(2-{[(9Z)-hexadec-9-enoyl]amino}ethyl)-beta-alaninamide, ... | | Authors: | Mindrebo, J.T, Chen, A, Noel, J.P, Burkart, M.D. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8FOL
 
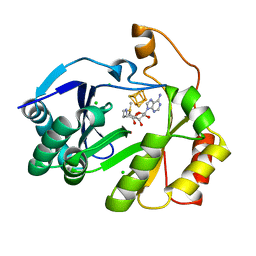 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM, alternate crystal form | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-31 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FO0
 
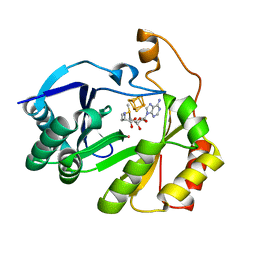 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to a partially cleaved SAM molecule | | Descriptor: | IRON/SULFUR CLUSTER, POTASSIUM ION, Pyruvate formate-lyase 1-activating enzyme, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-29 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FSI
 
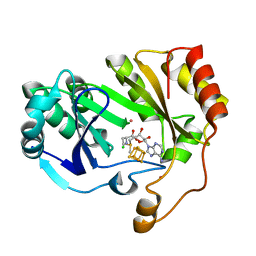 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Moody, J.D, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
1YPP
 
 | | ACID ANHYDRIDE HYDROLASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Harutyunyan, E.H, Kuranova, I.P, Lamzin, V.S, Dauter, Z, Wilson, K.S. | | Deposit date: | 1996-05-29 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of yeast inorganic pyrophosphatase complexed with manganese and phosphate.
Eur.J.Biochem., 239, 1996
|
|
4UT6
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 B7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 B7, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UTA
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C8 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8 HEAVY CHAIN, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8 LIGHT CHAIN, ENVELOPE GLYCOPROTEIN E, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UT9
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein dimer in complex with the ScFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C10, ENVELOPE GLYCOPROTEIN E | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UT7
 
 | | CRYSTAL STRUCTURE OF THE SCFV FRAGMENT OF THE BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4V1V
 
 | | Heterocyclase in complex with substrate and Cofactor | | Descriptor: | LYND, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Koehnke, J, Naismith, J.H. | | Deposit date: | 2014-10-02 | | Release date: | 2015-01-14 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Analysis of Leader Peptide Binding Enables Leader-Free Cyanobactin Processing.
Nat.Chem.Biol., 11, 2015
|
|
4UTB
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 A11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UTC
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN E, ... | | Authors: | Kikuti, C, Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4WSJ
 
 | | Crystal structure of a bacterial fucodiase in complex with 1-((1R,2R,3R,4R,5R,6R)-2,3,4-trihydroxy-5-methyl-7-azabicyclo[4.1.0]heptan-7-yl)ethan-1-one | | Descriptor: | Alpha-L-fucosidase, N-[(1S,2R,3R,4S,5R)-3,4,5-trihydroxy-2-methylcyclohexyl]acetamide, SULFATE ION | | Authors: | Davies, G.J, Wright, D.W. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | In vitroandin vivocomparative and competitive activity-based protein profiling of GH29 alpha-l-fucosidases.
Chem Sci, 6, 2015
|
|
4QWT
 
 | |
4RCH
 
 | | Discovery of 2-Pyridyl Ureas as Glucokinase Activators | | Descriptor: | 1-{3-[(2-ethylpyridin-3-yl)oxy]-5-(pyridin-2-ylsulfanyl)pyridin-2-yl}-3-methylurea, Glucokinase, alpha-D-glucopyranose | | Authors: | Voegtli, W, Vigers, G.P.A. | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of 2-pyridylureas as glucokinase activators.
J.Med.Chem., 57, 2014
|
|
7OSB
 
 | | Crystal Structure of a Double Mutant PETase (S238F/W159H) from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ethylene terephthalate) hydrolase, ... | | Authors: | Shakespeare, T.J, Zahn, M, Allen, M.D, McGeehan, J.E. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparative Performance of PETase as a Function of Reaction Conditions, Substrate Properties, and Product Accumulation.
ChemSusChem, 15, 2022
|
|
7P5F
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | Descriptor: | 5-cyclopropyl-1-[3-[3-(dimethylcarbamoyl)phenyl]phenyl]pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P5N
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | Descriptor: | 1-[3-[(1~{R},3~{S})-3-[(2~{R})-2-butylpyrrolidin-1-yl]carbonylcyclohexyl]phenyl]-5-cyclopropyl-pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P5P
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | Descriptor: | 5-[(1~{R},2~{R})-2-(1-methyl-1,2,3-triazol-4-yl)cyclopropyl]-1-[3-[3-[(2~{R})-2-propylpiperidin-1-yl]carbonylphenyl]phenyl]pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Davies, T.G. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P5E
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | Descriptor: | 1-[6-[3-(dimethylcarbamoyl)phenyl]pyridin-2-yl]-5-(trifluoromethyl)pyrazole-4-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Davies, T.G, Cleasby, A. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P5K
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-fluoranyl-3-[(2~{R})-2-propylpiperidin-1-yl]carbonyl-phenyl]phenyl]pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P58
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | Descriptor: | 1-[6-(3-propan-2-yloxyphenyl)pyridin-2-yl]-5-(trifluoromethyl)pyrazole-4-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Davies, T.G. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P5I
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | Descriptor: | 1-[3-[3-(dimethylcarbamoyl)phenyl]phenyl]-5-[(1~{R},2~{R})-2-(1-methyl-1,2,3-triazol-4-yl)cyclopropyl]pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Davies, T.G, Cleasby, A. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
4V1U
 
 | |
1AZC
 
 | | STRUCTURE OF APO-AZURIN FROM ALCALIGENES DENITRIFICANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION, SULFATE ION | | Authors: | Baker, E.N, Shepard, W.E.B, Kingston, R.L. | | Deposit date: | 1992-12-16 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of apo-azurin from Alcaligenes denitrificans at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
