1AJ8
 
 | | CITRATE SYNTHASE FROM PYROCOCCUS FURIOSUS | | Descriptor: | CITRATE SYNTHASE, CITRIC ACID, COENZYME A | | Authors: | Russell, R.J.M, Ferguson, J.M.C, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 1997-05-16 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of citrate synthase from the hyperthermophilic archaeon pyrococcus furiosus at 1.9 A resolution,.
Biochemistry, 36, 1997
|
|
1A59
 
 | | COLD-ACTIVE CITRATE SYNTHASE | | Descriptor: | CITRATE SYNTHASE, CITRIC ACID, COENZYME A | | Authors: | Russell, R.J.M, Gerike, U, Danson, M.J, Hough, D.W, Taylor, G.L. | | Deposit date: | 1998-02-20 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium.
Structure, 6, 1998
|
|
4GAY
 
 | | Structure of the broadly neutralizing antibody AP33 | | Descriptor: | NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, NEUTRALIZING ANTIBODY AP33 LIGHT CHAIN, TRIETHYLENE GLYCOL | | Authors: | Potter, J.A, Owsianka, A, Jeffery, N, Matthews, D, Keck, Z, Lau, P, Foung, S.K.H, Taylor, G.L, Patel, A.H. | | Deposit date: | 2012-07-26 | | Release date: | 2012-10-10 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
4GAJ
 
 | | Structure of the broadly neutralizing antibody AP33 in complex with its HCV epitope (E2 residues 411-424) | | Descriptor: | Genome polyprotein, NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, NEUTRALIZING ANTIBODY AP33 LIGHT CHAIN | | Authors: | Potter, J.A, Owsianka, A, Taylor, G.L, Patel, A.H. | | Deposit date: | 2012-07-25 | | Release date: | 2012-10-10 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
4GAG
 
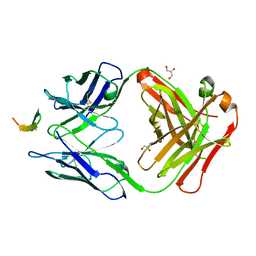 | | Structure of the broadly neutralizing antibody AP33 in complex with its HCV epitope (E2 residues 412-423) | | Descriptor: | GLYCEROL, Genome polyprotein, NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, ... | | Authors: | Potter, J.A, Owsianka, A, Angus, A.G.N, Jeffery, N, Matthews, D, Keck, Z, Lau, P, Foung, S.K.H, Taylor, G.L, Patel, A.H. | | Deposit date: | 2012-07-25 | | Release date: | 2012-10-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
1W3T
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with D-KDGal, D-Glyceraldehyde and pyruvate | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, 3-DEOXY-D-LYXO-HEXONIC ACID, D-Glyceraldehyde, ... | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-19 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
1W3N
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with D-KDG | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, 3-DEOXY-D-ARABINO-HEXONIC ACID, GLYCEROL | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-17 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
1W3I
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with pyruvate | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, GLYCEROL, PYRUVIC ACID | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-15 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
2X7B
 
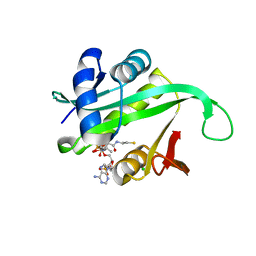 | | Crystal structure of the N-terminal acetylase Ard1 from Sulfolobus solfataricus P2 | | Descriptor: | CHLORIDE ION, COENZYME A, N-ACETYLTRANSFERASE SSO0209 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Mackay, D, White, M.F, Taylor, G.L, Naismith, J.H. | | Deposit date: | 2010-02-25 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
1O7X
 
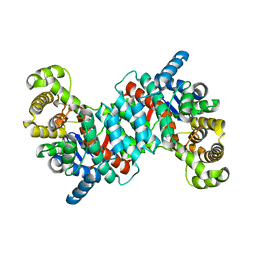 | | Citrate synthase from Sulfolobus solfataricus | | Descriptor: | CITRATE SYNTHASE | | Authors: | Bell, G.S, Russell, R.J.M, Connaris, H, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Stepwise Adaptations of Citrate Synthase to Survival at Life'S Extremes. From Psychrophile to Hyperthermophile.
Eur.J.Biochem., 269, 2002
|
|
2VAR
 
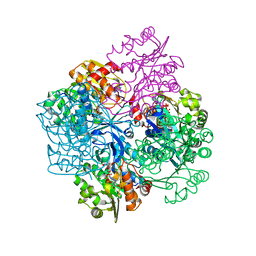 | | Crystal structure of Sulfolobus solfataricus 2-keto-3-deoxygluconate kinase complexed with 2-keto-3-deoxygluconate | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, 3-deoxy-alpha-D-erythro-hex-2-ulofuranosonic acid, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Potter, J.A, Theodossis, A, Kerou, M, Lamble, H.J, Bull, S.D, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2007-09-04 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Sulfolobus Solfataricus 2-Keto-3-Deoxygluconate Kinase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2VK7
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, EXO-ALPHA-SIALIDASE | | Authors: | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
2VGQ
 
 | | Crystal Structure of Human IPS-1 CARD | | Descriptor: | SULFATE ION, Sugar ABC transporter substrate-binding protein,Mitochondrial antiviral-signaling protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Potter, J.A, Randall, R.E, Taylor, G.L. | | Deposit date: | 2007-11-15 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Ips-1 Caspase Activation Recruitment Domain
Bmc Struct.Biol., 8, 2008
|
|
2VK6
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, EXO-ALPHA-SIALIDASE, ... | | Authors: | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
2VK5
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | Descriptor: | CALCIUM ION, EXO-ALPHA-SIALIDASE, GLYCEROL | | Authors: | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
2V1L
 
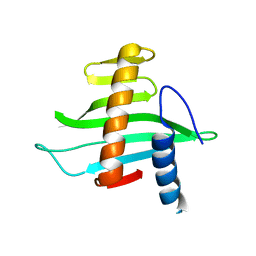 | | Structure of the conserved hypothetical protein VC1805 from pathogenicity island VPI-2 of Vibrio cholerae O1 biovar eltor str. N16961 shares structural homology with the human P32 protein | | Descriptor: | HYPOTHETICAL PROTEIN | | Authors: | Sheikh, M.A, Potter, J.A, Johnson, K.A, Boyd, E.F, Taylor, G.L. | | Deposit date: | 2007-05-25 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Vc1805, a Conserved Hypothetical Protein from a Vibrio Cholerae Pathogenicity Island, Reveals Homology to Human P32.
Proteins, 71, 2008
|
|
2V78
 
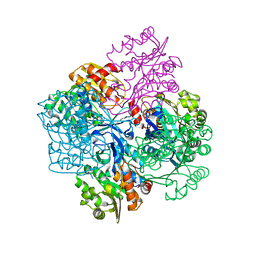 | | Crystal structure of Sulfolobus solfataricus 2-keto-3-deoxygluconate kinase | | Descriptor: | FRUCTOKINASE | | Authors: | Potter, J.A, Theodossis, A, Kerou, M, Lamble, H.J, Bull, S.D, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Sulfolobus Solfataricus 2-Keto-3-Deoxygluconate Kinase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2VW1
 
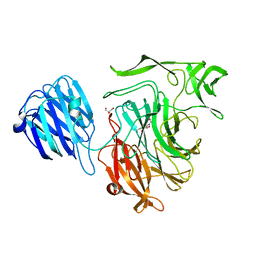 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
2W68
 
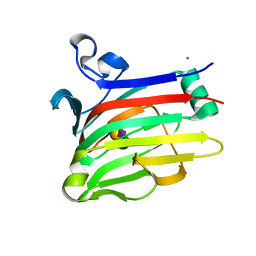 | |
2W38
 
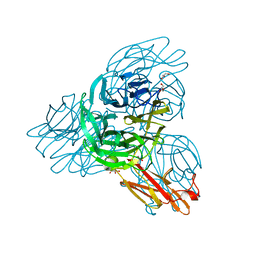 | | Crystal structure of the pseudaminidase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, SIALIDASE | | Authors: | Xu, G, Ryan, C, Kiefel, M.J, Wilson, J.C, Taylor, G.L. | | Deposit date: | 2008-11-07 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on the Pseudomonas Aeruginosa Sialidase-Like Enzyme Pa2794 Suggest Substrate and Mechanistic Variations.
J.Mol.Biol., 386, 2009
|
|
2VVZ
 
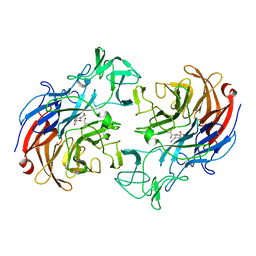 | | Structure of the catalytic domain of Streptococcus pneumoniae sialidase NanA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CHLORIDE ION, SIALIDASE A | | Authors: | Xu, G, Li, X, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Catalytic Domain of Streptococcus Pneumoniae Sialidase Nana.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2W57
 
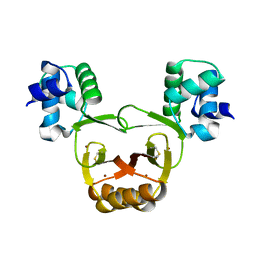 | |
2W56
 
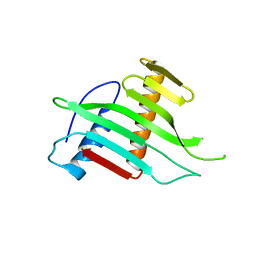 | |
2VW2
 
 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
2VW0
 
 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
