1UDY
 
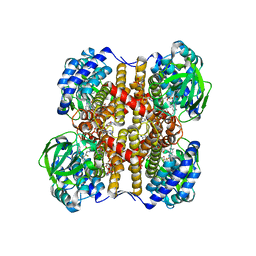 | | Medium-Chain Acyl-CoA Dehydrogenase with 3-Thiaoctanoyl-CoA | | Descriptor: | 3-THIAOCTANOYL-COENZYME A, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Satoh, A, Nakajima, Y, Miyahara, I, Hirotsu, K, Tanaka, T, Nishina, Y, Shiga, K, Tamaoki, H, Setoyama, C, Miura, R. | | Deposit date: | 2003-05-07 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the transition state analog of medium-chain acyl-CoA dehydrogenase. Crystallographic and molecular orbital studies on the charge-transfer complex of medium-chain acyl-CoA dehydrogenase with 3-thiaoctanoyl-CoA
J.BIOCHEM.(TOKYO), 134, 2003
|
|
2DI2
 
 | | NMR structure of the HIV-2 nucleocapsid protein | | Descriptor: | Nucleocapsid protein p7, ZINC ION | | Authors: | Matsui, T, Kodera, Y, Endoh, H, Miyauchi, E, Komatsu, H, Sato, K, Tanaka, T, Kohno, T, Maeda, T. | | Deposit date: | 2006-03-27 | | Release date: | 2007-03-13 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | RNA Recognition Mechanism of the Minimal Active Domain of the Human Immunodeficiency Virus Type-2 Nucleocapsid Protein
J.Biochem.(Tokyo), 141, 2007
|
|
2DWN
 
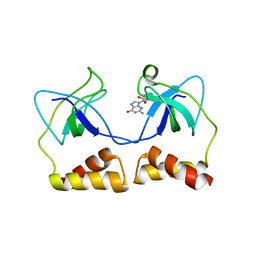 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | DNA (5'-D(*A*G)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWM
 
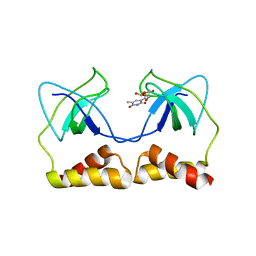 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*T)-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWL
 
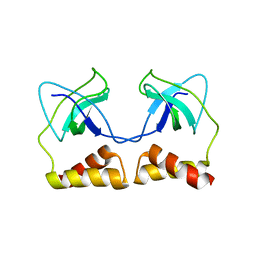 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*(DC))-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
5D48
 
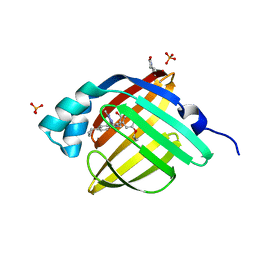 | | Crystal Structure of FABP4 in complex with 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy) phenyl]-1H-indol-1-yl}propanoic acid | | Descriptor: | 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy)phenyl]-1H-indol-1-yl}propanoic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D47
 
 | | Crystal Structure of FABP4 in complex with 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl] propanoic acid | | Descriptor: | 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl]propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
3AQP
 
 | | Crystal structure of SecDF, a translocon-associated membrane protein, from Thermus thrmophilus | | Descriptor: | Probable SecDF protein-export membrane protein | | Authors: | Tsukazaki, T, Mori, H, Echizen, Y, Ishitani, R, Fukai, S, Tanaka, T, Perederina, A, Vassylyev, D.G, Kohno, T, Ito, K, Nureki, O. | | Deposit date: | 2010-11-16 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of a membrane component SecDF that enhances protein export
Nature, 474, 2011
|
|
2ZMH
 
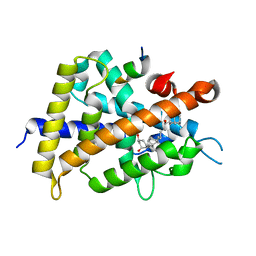 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1R,2E,4R)-4-hydroxy-1-methyl-4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]but-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-18 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
2ZMJ
 
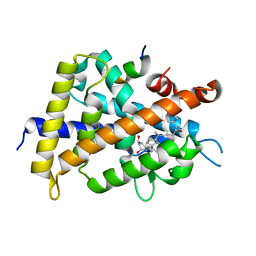 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1S,2E,5R)-5-hydroxy-1-methyl-6-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hex-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
2ZMI
 
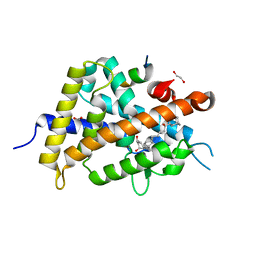 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1S,2E,5R)-5-hydroxy-1-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]pent-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
2YR2
 
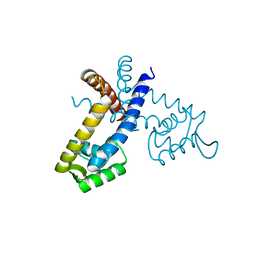 | |
3AA9
 
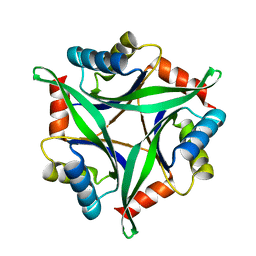 | | Crystal Structure Analysis of the Mutant CutA1 (E61V) from E. coli | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2009-11-12 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
2ZZA
 
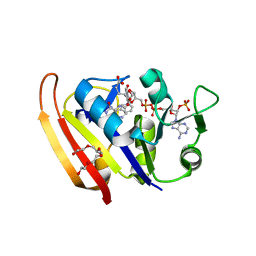 | | Moritella profunda Dihydrofolate reductase complex with NADP+ and Folate | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hata, K, Tanaka, T, Murakami, C, Ohmae, E, Gekko, K, Shiro, Y, Akasaka, K. | | Deposit date: | 2009-02-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Moritella profunda Dihydrofolate reductase complex with NADP+ and Folate
To be Published
|
|
2ZCB
 
 | | Crystal Structure of ubiquitin P37A/P38A | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Kitahara, R, Tanaka, T, Sakata, E, Yamaguchi, Y, Kato, K, Yokoyama, S. | | Deposit date: | 2007-11-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of ubiquitin P37A/P38A
To be published
|
|
3AH6
 
 | | Remarkable improvement of the heat stability of CutA1 from E.coli by rational protein designing | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
3A5T
 
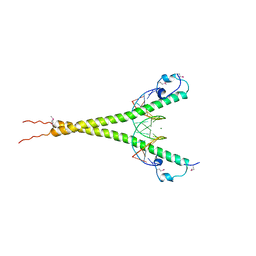 | | Crystal structure of MafG-DNA complex | | Descriptor: | 5'-D(*CP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*G)-3', MAGNESIUM ION, ... | | Authors: | Kurokawa, H, Motohashi, H, Sueno, S, Kimura, M, Takagawa, H, Kanno, Y, Yamamoto, M, Tanaka, T. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-13 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Alternative DNA Recognition by Maf Transcription Factors
Mol.Cell.Biol., 29, 2009
|
|
2ZCC
 
 | | Ubiquitin crystallized under high pressure | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Kitahara, R, Tanaka, T, Yamashita, M, Araya, K, Yokoyama, S, Akasaka, K, Taniguchi, Y, Kato, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Ubiquitin crystallized under high pressure
to be published
|
|
3AA8
 
 | | Crystal Structure Analysis of the Mutant CutA1 (S11V/E61V) from E. coli | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2009-11-12 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
3VNG
 
 | | Crystal Structure of Keap1 in Complex with Synthetic Small Molecular based on a co-crystallization | | Descriptor: | 2-(3-((3-(5-(furan-2-yl)-1,3,4-oxadiazol-2-yl)ureido)methyl)phenoxy)acetic acid, Kelch-like ECH-associated protein 1 | | Authors: | Kunishima, N, Tanaka, T, Satoh, M, Saburi, H. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Keap1 in Complex with Synthetic Small Molecular based on a co-crystallization
To be Published
|
|
3W59
 
 | |
3WN8
 
 | | Crystal Structure of Collagen-Model Peptide, (POG)3-PRG-(POG)4 | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Haga, M, Noguchi, K, Tanaka, T. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Preferred side-chain conformation of arginine residues in a triple-helical structure.
Biopolymers, 101, 2014
|
|
3W58
 
 | |
3VNH
 
 | | Crystal Structure of Keap1 Soaked with Synthetic Small Molecular | | Descriptor: | 2-(3-((3-(5-(furan-2-yl)-1,3,4-oxadiazol-2-yl)ureido)methyl)phenoxy)acetic acid, Kelch-like ECH-associated protein 1 | | Authors: | Kunishima, N, Tanaka, T, Satoh, M, Saburi, H. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Keap1 Soaked with Synthetic Small Molecular
To be Published
|
|
