1VQH
 
 | |
1VQE
 
 | |
1VQG
 
 | |
1VQA
 
 | |
1VQJ
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 (V35I) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VQF
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 AND ILE 47 REPLACED BY VAL 47 (V35I, I47V) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VQC
 
 | |
1VQI
 
 | | GENE V PROTEIN MUTANT WITH ILE 47 REPLACED BY VAL 47 (I47V) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VQD
 
 | |
1WDC
 
 | | SCALLOP MYOSIN REGULATORY DOMAIN | | Descriptor: | CALCIUM ION, MAGNESIUM ION, SCALLOP MYOSIN | | Authors: | Houdusse, A, Cohen, C. | | Deposit date: | 1996-01-19 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the regulatory domain of scallop myosin at 2 A resolution: implications for regulation.
Structure, 4, 1996
|
|
1YHB
 
 | | CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX | | Descriptor: | GENE V PROTEIN | | Authors: | Guan, Y, Zhang, H, Konings, R.N.H, Hilbers, C.W, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1994-04-14 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Y41H and Y41F mutants of gene V protein from Ff phage suggest possible protein-protein interactions in the GVP-ssDNA complex.
Biochemistry, 33, 1994
|
|
1YHA
 
 | | CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX | | Descriptor: | GENE V PROTEIN | | Authors: | Guan, Y, Zhang, H, Konings, R.N.H, Hilbers, C.W, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1994-04-14 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Y41H and Y41F mutants of gene V protein from Ff phage suggest possible protein-protein interactions in the GVP-ssDNA complex.
Biochemistry, 33, 1994
|
|
1MYZ
 
 | | CO COMPLEX OF MYOGLOBIN MB-YQR AT RT SOLVED FROM LAUE DATA. | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bourgeois, D, Vallone, B, Schotte, F, Arcovito, A, Miele, A.E, Sciara, G, Wulff, M, Anfinrud, P, Brunori, M. | | Deposit date: | 2002-10-04 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complex landscape of protein
structural dynamics unveiled by
nanosecond Laue crystallography.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1MZL
 
 | | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Shin, D.H, Lee, J.Y, Hwang, K.Y, Kim, K.K, Suh, S.W. | | Deposit date: | 1995-01-26 | | Release date: | 1996-08-01 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings.
Structure, 3, 1995
|
|
1MZ0
 
 | | STRUCTURE OF MYOGLOBIN MB-YQR 316 ns AFTER PHOTOLYSIS OF CARBON MONOXIDE SOLVED FROM LAUE DATA AT RT. | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bourgeois, D, Vallone, B, Schotte, F, Arcovito, A, Miele, A.E, Sciara, G, Wulff, M, Anfinrud, P, Brunori, M. | | Deposit date: | 2002-10-04 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complex landscape of protein structural dynamics unveiled by
nanosecond Laue crystallography.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
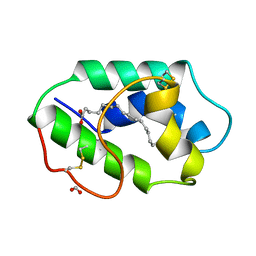
1MZM
 
 | |
1O76
 
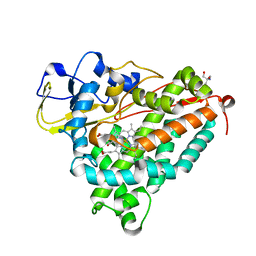 | | CYANIDE COMPLEX OF P450CAM FROM PSEUDOMONAS PUTIDA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYANIDE ION, ... | | Authors: | Fedorov, R, Ghosh, D, Schlichting, I. | | Deposit date: | 2002-10-23 | | Release date: | 2002-12-19 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Cyanide Complexes of P450Cam and the Oxygenase Domain of Inducible Nitric Oxide Synthase-Structural Models of the Short-Lived Oxygen Complexes
Arch.Biochem.Biophys., 409, 2003
|
|
1OB0
 
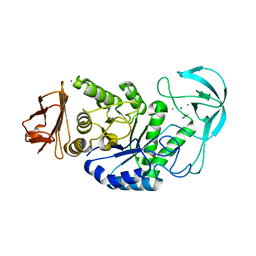 | | Kinetic stabilization of Bacillus licheniformis alpha-amylase through introduction of hydrophobic residues at the surface | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 2003-01-21 | | Release date: | 2003-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Kinetic Stabilization of Bacillus Licheniformis Alpha-Amylase Through Introduction of Hydrophobic Residues at the Surface
J.Biol.Chem., 278, 2003
|
|
1OIL
 
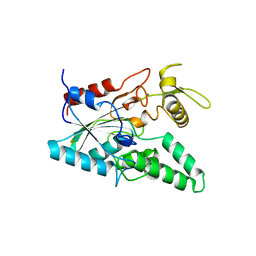 | | STRUCTURE OF LIPASE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Kim, K.K, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 1996-12-06 | | Release date: | 1997-05-15 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor.
Structure, 5, 1997
|
|
1RLD
 
 | | SOLID-STATE PHASE TRANSITION IN THE CRYSTAL STRUCTURE OF RIBULOSE 1,5-BIPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE | | Descriptor: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Eisenberg, D. | | Deposit date: | 1993-12-10 | | Release date: | 1994-04-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solid-state phase transition in the crystal structure of ribulose 1,5-bisphosphate carboxylase/oxygenase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1RLC
 
 | | CRYSTAL STRUCTURE OF THE UNACTIVATED RIBULOSE 1, 5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE COMPLEXED WITH A TRANSITION STATE ANALOG, 2-CARBOXY-D-ARABINITOL 1,5-BISPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Cascio, D, Eisenberg, D. | | Deposit date: | 1993-08-04 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the unactivated ribulose 1,5-bisphosphate carboxylase/oxygenase complexed with a transition state analog, 2-carboxy-D-arabinitol 1,5-bisphosphate.
Protein Sci., 3, 1994
|
|
1RZL
 
 | | RICE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, NONSPECIFIC LIPID TRANSFER PROTEIN, SULFATE ION | | Authors: | Lee, J.Y, Min, K.S, Cha, H, Shin, D.H, Hwang, K.Y, Suh, S.W. | | Deposit date: | 1997-10-09 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rice non-specific lipid transfer protein: the 1.6 A crystal structure in the unliganded state reveals a small hydrophobic cavity.
J.Mol.Biol., 276, 1998
|
|
1J7U
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa AMPPNP Complex | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
1J7L
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa ADP Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, MAGNESIUM ION | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-17 | | Release date: | 2001-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
1J7I
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa Apoenzyme | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
