4JCS
 
 | | Crystal structure of Enoyl-CoA hydratase/isomerase from Cupriavidus metallidurans CH34 | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Eswaramoorthy, S, Chamala, S, Chamala, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Enoyl-CoA hydratase/isomerase from Cupriavidus metallidurans CH34
To be Published
|
|
4K29
 
 | | Crystal structure of an enoyl-CoA hydratase/isomerase from Xanthobacter autotrophicus Py2 | | Descriptor: | Enoyl-CoA hydratase/isomerase, GLYCEROL, L(+)-TARTARIC ACID | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-08 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/isomerase from Xanthobacter autotrophicus Py2
TO BE PUBLISHED
|
|
4EW6
 
 | |
4K2N
 
 | | Crystal structure of an enoyl-CoA hydratase/ carnithine racemase from Magnetospirillum magneticum | | Descriptor: | Enoyl-CoA hydratase/carnithine racemase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-09 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/ carnithine racemase from Magnetospirillum magneticum
To be Published
|
|
4K3W
 
 | | Crystal structure of an enoyl-CoA hydratase/isomerase from Marinobacter aquaeolei | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-11 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/isomerase from Marinobacter aquaeolei
To be Published
|
|
4FB5
 
 | |
4MYM
 
 | | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, AL Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-27 | | Release date: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides.
To be Published
|
|
4G9Q
 
 | | Crystal structure of a 4-carboxymuconolactone decarboxylase | | Descriptor: | 4-carboxymuconolactone decarboxylase | | Authors: | Hickey, H.D, Mcgillick, B.E, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-24 | | Release date: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a 4-carboxymuconolactone decarboxylase
To be Published
|
|
2POF
 
 | |
2Q09
 
 | | Crystal structure of Imidazolonepropionase from environmental sample with bound inhibitor 3-(2,5-Dioxo-imidazolidin-4-yl)-propionic acid | | Descriptor: | 3-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4-YL]PROPANOIC ACID, FE (III) ION, Imidazolonepropionase | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A common catalytic mechanism for proteins of the HutI family.
Biochemistry, 47, 2008
|
|
2QQ6
 
 | |
2QVG
 
 | |
2RK9
 
 | | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase superfamily member from Vibrio splendidus 12B01 | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Tyagi, R, Eswaramoorthy, S, Sauder, J.M, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase superfamily member from Vibrio splendidus 12B01.
To be Published
|
|
4ZJX
 
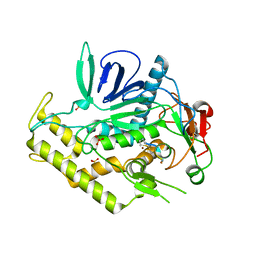 | |
1F1M
 
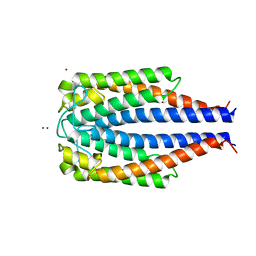 | | CRYSTAL STRUCTURE OF OUTER SURFACE PROTEIN C (OSPC) | | Descriptor: | OUTER SURFACE PROTEIN C, ZINC ION | | Authors: | Kumaran, D, Eswaramoorthy, S, Dunn, J.J, Swaminathan, S. | | Deposit date: | 2000-05-19 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.
EMBO J., 20, 2001
|
|
4DUP
 
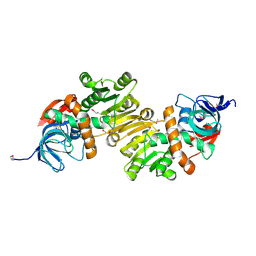 | | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42 | | Descriptor: | quinone oxidoreductase | | Authors: | Kumaran, D, Rice, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
4E21
 
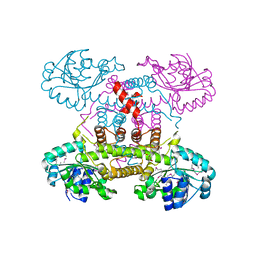 | | The crystal structure of 6-phosphogluconate dehydrogenase from Geobacter metallireducens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 6-phosphogluconate dehydrogenase (Decarboxylating) | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-07 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | The crystal structure of 6-phosphogluconate dehydrogenase from Geobacter metallireducens
To be Published
|
|
4HGV
 
 | | Crystal structure of a fumarate hydratase | | Descriptor: | Fumarate hydratase class II, SULFATE ION | | Authors: | Eswaramoorthy, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a fumarate hydratase
To be Published
|
|
2PUZ
 
 | | Crystal structure of Imidazolonepropionase from Agrobacterium tumefaciens with bound product N-formimino-L-Glutamate | | Descriptor: | CHLORIDE ION, FE (III) ION, Imidazolonepropionase, ... | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray structure of imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution.
Proteins, 69, 2007
|
|
4J2U
 
 | | Crystal structure of an enoyl-CoA hydratase from Rhodobacter sphaeroides 2.4.1 | | Descriptor: | Enoyl-CoA hydratase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-05 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase from Rhodobacter sphaeroides 2.4.1
To be Published
|
|
4JFC
 
 | | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666 | | Descriptor: | Enoyl-CoA hydratase, GLYCEROL | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666
To be Published
|
|
4JCU
 
 | | Crystal structure of a 5-carboxymethyl-2-hydroxymuconate isomerase from Deinococcus radiodurans R1 | | Descriptor: | 5-carboxymethyl-2-hydroxymuconate isomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a 5-carboxymethyl-2-hydroxymuconate isomerase from Deinococcus radiodurans R1
To be Published
|
|
4KPK
 
 | | Crystal structure of a enoyl-CoA hydratase from Shewanella pealeana ATCC 700345 | | Descriptor: | 1,2-ETHANEDIOL, Enoyl-CoA hydratase/isomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-05-13 | | Release date: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Shewanella pealeana ATCC 700345
To be Published
|
|
4KD6
 
 | | Crystal structure of a Enoyl-CoA hydratase/isomerase from Burkholderia graminis C4D1M | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a Enoyl-CoA hydratase/isomerase from Burkholderia graminis C4D1M
To be Published
|
|
4LK5
 
 | | Crystal structure of a enoyl-CoA hydratase from Mycobacterium avium subsp. paratuberculosis K-10 | | Descriptor: | enoyl-CoA hydratase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-06 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Mycobacterium avium subsp. paratuberculosis K-10
To be Published
|
|
