6BS9
 
 | | Stage III sporulation protein AB (SpoIIIAB) | | 分子名称: | SULFATE ION, Stage III sporulation protein AB | | 著者 | Strynadka, N.C.J, Zeytuni, N, Camp, A.H, Flanagan, K.A. | | 登録日 | 2017-12-01 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural characterization of SpoIIIAB sporulation-essential protein in Bacillus subtilis.
J. Struct. Biol., 202, 2018
|
|
6DCS
 
 | | Stage III sporulation protein AF (SpoIIIAF) | | 分子名称: | SULFATE ION, Stage III sporulation protein AF | | 著者 | Strynadka, N.C.J, Zeytuni, N, Camp, A.H, Flanagan, K.A. | | 登録日 | 2018-05-08 | | 公開日 | 2018-07-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and biochemical characterization of SpoIIIAF, a component of a sporulation-essential channel in Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
6ZTG
 
 | | Spor protein DedD | | 分子名称: | Cell division protein DedD | | 著者 | Pazos, M, Peters, K, Boes, A, Safaei, Y, Kenward, C, Caveney, N.A, Laguri, C, Breukink, E, Strynadka, N.C.J, Simorre, J.P, Terrak, M, Vollmer, W. | | 登録日 | 2020-07-20 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SPOR Proteins Are Required for Functionality of Class A Penicillin-Binding Proteins in Escherichia coli.
Mbio, 11, 2020
|
|
6Q16
 
 | | Focussed refinement of InvGN0N1:PrgHK:SpaPQR:PrgIJ from Salmonella SPI-1 injectisome NC-base | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-08-02 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-15 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6N7O
 
 | |
2DRJ
 
 | |
8SXR
 
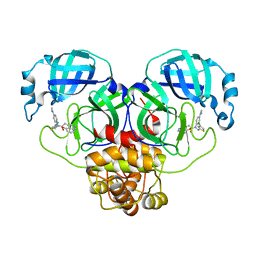 | | Crystal structure of SARS-CoV-2 Mpro with C5a | | 分子名称: | 3C-like proteinase nsp5, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(5-hydroxyisoquinolin-4-yl)acetamide | | 著者 | Worrall, L.J, Kenward, C, Lee, J, Strynadka, N.C.J. | | 登録日 | 2023-05-23 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.114 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8V33
 
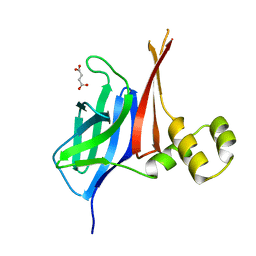 | |
8VA1
 
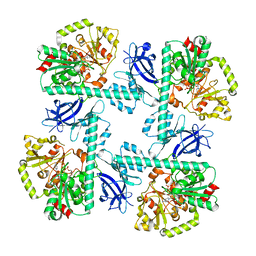 | |
8V34
 
 | |
2F2H
 
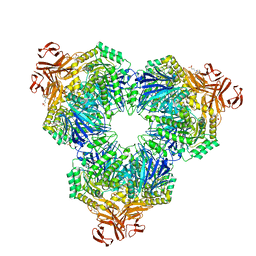 | | Structure of the YicI thiosugar Michaelis complex | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-NITROPHENYL 6-THIO-6-S-ALPHA-D-XYLOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, GLYCEROL, ... | | 著者 | Kim, Y.-W, Lovering, A.L, Strynadka, N.C.J, Withers, S.G. | | 登録日 | 2005-11-16 | | 公開日 | 2006-02-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Expanding the Thioglycoligase Strategy to the Synthesis of alpha-linked Thioglycosides Allows Structural Investigation of the Parent Enzyme/Substrate Complex
J.Am.Chem.Soc., 128, 2006
|
|
7TC5
 
 | | All Phe-Azurin variant - F15Y | | 分子名称: | Azurin, COPPER (II) ION, NITRATE ION, ... | | 著者 | Fedoretz-Maxwell, B.P, Worrall, L.J, Strynadka, N.C.J, Warren, J.J. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins.
Inorg.Chem., 61, 2022
|
|
7TC6
 
 | | All Phe-Azurin variant - F15W | | 分子名称: | Azurin, COPPER (II) ION, NITRATE ION | | 著者 | Fedoretz-Maxwell, B.P, Worrall, L.J, Strynadka, N.C.J, Warren, J.J. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins.
Inorg.Chem., 61, 2022
|
|
7UZ2
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-fluoro-valienide. | | 分子名称: | (1R,2S,3R,4R)-5-fluoro-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Beta-galactosidase | | 著者 | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | 登録日 | 2022-05-08 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7UZ1
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-bromo-valienide. | | 分子名称: | (1R,2S,3R,4R)-5-bromo-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Beta-galactosidase | | 著者 | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Karimi, R, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | 登録日 | 2022-05-08 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8EXQ
 
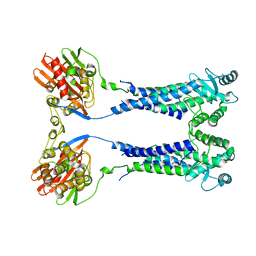 | | Cryo-EM structure of S. aureus BlaR1 with C1 symmetry | | 分子名称: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | 著者 | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | 登録日 | 2022-10-25 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-01-25 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXR
 
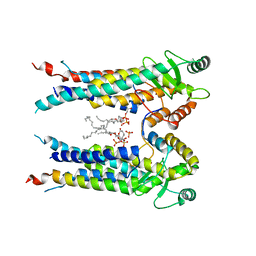 | | Cryo-EM structure of S. aureus BlaR1 TM and zinc metalloprotease domain | | 分子名称: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Beta-lactam sensor/signal transducer BlaR1, PHOSPHATE ION, ... | | 著者 | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | 登録日 | 2022-10-25 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-01-25 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXS
 
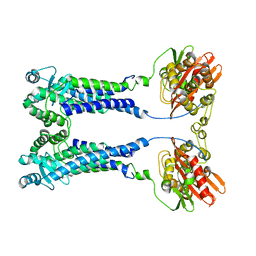 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant | | 分子名称: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | 著者 | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2022-10-25 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-01-25 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXP
 
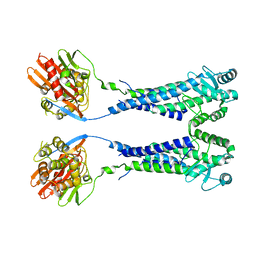 | | Cryo-EM structure of S. aureus BlaR1 with C2 symmetry | | 分子名称: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | 著者 | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | 登録日 | 2022-10-25 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-01-25 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXT
 
 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant in complex with ampicillin | | 分子名称: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | 著者 | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2022-10-25 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-01-25 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8FFJ
 
 | | Structure of Zanidatamab bound to HER2 | | 分子名称: | Receptor tyrosine-protein kinase erbB-2, Zanidatamab Heavy Chain A, Zanidatamab Heavy Chain B, ... | | 著者 | Worrall, L.J, Atkinson, C.E, Sanches, M, Dixit, S, Strynadka, N.C.J. | | 登録日 | 2022-12-08 | | 公開日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (7.5 Å) | | 主引用文献 | Zanidatamab, an Anti-HER2 Biparatopic that Induces Unique Surface HER2 Clusters and Complement-Dependent Cytotoxicity
To be Published
|
|
1KN9
 
 | |
6C39
 
 | |
6C3K
 
 | |
6CO9
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB COCHEA-COA complex | | 分子名称: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | 著者 | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | 登録日 | 2018-03-12 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
