6COJ
 
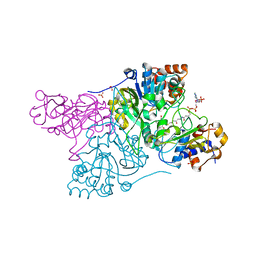 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB E105A COCHEA-COA complex | | Descriptor: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CB2
 
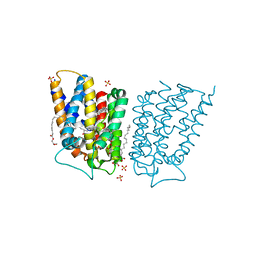 | | Crystal structure of Escherichia coli UppP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Undecaprenyl-diphosphatase | | Authors: | Workman, S.D, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2018-02-01 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling.
Nat Commun, 9, 2018
|
|
6CON
 
 | | Crystal structure of Mycobacterium tuberculosis IpdAB | | Descriptor: | CoA-transferase subunit alpha, CoA-transferase subunit beta | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CO6
 
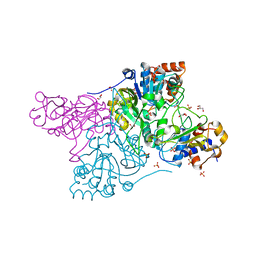 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB | | Descriptor: | GLYCEROL, Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DWB
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome needle filament | | Descriptor: | Protein PrgI | | Authors: | Hu, J, Hong, C, Worrall, L.J, Vuckovic, M, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DV3
 
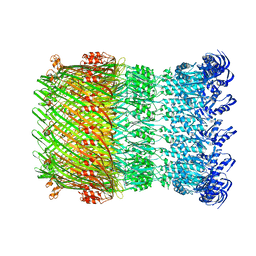 | | Structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG in the open gate state | | Descriptor: | Protein InvG | | Authors: | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DFE
 
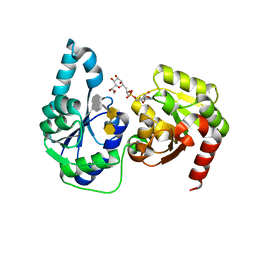 | | The structure of a ternary complex of E. coli WaaC | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-acetamido-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, ADP-heptose--LPS heptosyltransferase, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R)-1-{(2S,3S,4R,5S,6R)-6-[(1S)-1,2-dihydroxyethyl]-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl}propan-2-yl hydrogen (R)-phosphate (non-preferred name) | | Authors: | Worrall, L.J, Blaukopf, M, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2018-05-14 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Insights into Heptosyltransferase I Catalysis and Inhibition through the Structure of Its Ternary Complex.
Structure, 26, 2018
|
|
6DV6
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG (residues 176-end) in the open gate state | | Descriptor: | Protein InvG | | Authors: | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DUZ
 
 | | Structure of the periplasmic domains of PrgH and PrgK from the assembled Salmonella type III secretion injectisome needle complex | | Descriptor: | Lipoprotein PrgK, Protein PrgH | | Authors: | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DZ8
 
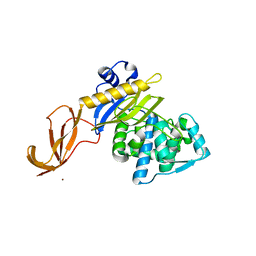 | |
6UEX
 
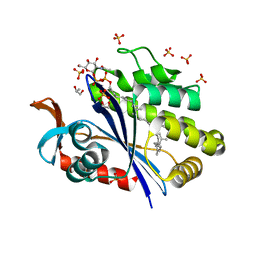 | | Crystal structure of S. aureus LcpA in complex with octaprenyl-pyrophosphate-GlcNAc | | Descriptor: | 2-(acetylamino)-2-deoxy-1-O-[(S)-hydroxy{[(S)-hydroxy{[(2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl]oxy}phosphoryl]oxy}phosphoryl]-alpha-D-glucopyranose, GLYCEROL, Regulatory protein MsrR, ... | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
6UF6
 
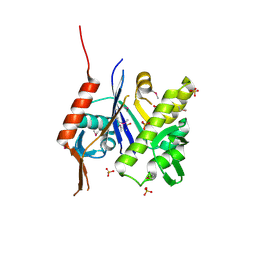 | | Crystal structure of B. subtilis TagU | | Descriptor: | GLYCEROL, Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagU, SULFATE ION | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-29 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
6UF3
 
 | | Crystal structure of B. subtilis TagV | | Descriptor: | Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagV | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
6UF5
 
 | | Crystal structure of B. subtilis TagT | | Descriptor: | Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagT | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
6U2D
 
 | | PmtCD peptide exporter basket domain | | Descriptor: | ABC transporter ATP-binding protein, IODIDE ION, SULFATE ION | | Authors: | Zeytuni, N, Strynadka, N.C.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
6W5Q
 
 | | Structure of the globular C-terminal domain of P. aeruginosa LpoP | | Descriptor: | Peptidoglycan synthase activator LpoP, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Caveney, N.A, Robb, C.S, Simorre, J.P, Strynadka, N.C.J. | | Deposit date: | 2020-03-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Peptidoglycan Synthase Activator LpoP in Pseudomonas aeruginosa.
Structure, 28, 2020
|
|
6XFK
 
 | | Crystal structure of the type III secretion system pilotin-secretin complex InvH-InvG | | Descriptor: | SULFATE ION, Type 3 secretion system pilotin, Type 3 secretion system secretin | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
6XFU
 
 | |
6NJP
 
 | | Structure of the assembled ATPase EscN in complex with its central stalk EscO from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, EscO, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
6NJO
 
 | | Structure of the assembled ATPase EscN from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
6NTZ
 
 | | Crystal structure of E. coli PBP5-meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, D-alanyl-D-alanine carboxypeptidase | | Authors: | Caveney, N.A, Strynadka, N.C.J, Caballero, G, Worrall, L.J. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli.
Nat Commun, 10, 2019
|
|
6NTW
 
 | | Crystal structure of E. coli YcbB | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Probable L,D-transpeptidase YcbB, SULFATE ION | | Authors: | Caveney, N.A, Strynadka, N.C.J, Caballero, G, Worrall, L.J. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-20 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli.
Nat Commun, 10, 2019
|
|
6O9S
 
 | |
6O9W
 
 | |
6PEP
 
 | | Focussed refinement of InvGN0N1:SpaPQR:PrgIJ from the Salmonella SPI-1 injectisome needle complex | | Descriptor: | Protein InvG, Protein PrgH, Protein PrgI, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-06-20 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
