1MNQ
 
 | | Thioesterase Domain of Picromycin Polyketide Synthase (PICS TE), pH 8.4 | | Descriptor: | polyketide synthase IV | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
1MO2
 
 | | Thioesterase Domain from 6-Deoxyerythronolide Synthase (DEBS TE), pH 8.5 | | Descriptor: | Erythronolide synthase, modules 5 and 6 | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
1MN6
 
 | | Thioesterase Domain from Picromycin Polyketide Synthase, pH 7.6 | | Descriptor: | polyketide synthase IV | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
2NQP
 
 | |
2NS1
 
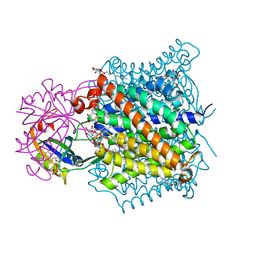 | | Crystal structure of the e. coli ammonia channel AMTB complexed with the signal transduction protein GLNK | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Ammonia channel, ... | | Authors: | Gruswitz, F, O'Connell III, J, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2006-11-02 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2NRE
 
 | |
2O9F
 
 | |
2O9E
 
 | |
2O9D
 
 | |
2O9G
 
 | |
2NR0
 
 | |
2OML
 
 | | crystal structure of E. coli pseudouridine synthase RluE | | Descriptor: | Ribosomal large subunit pseudouridine synthase E, SULFATE ION | | Authors: | Pan, H, Ho, J.D, Stroud, R.M, Finer-Moore, J. | | Deposit date: | 2007-01-22 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Crystal Structure of E. coli rRNA Pseudouridine Synthase RluE.
J.Mol.Biol., 367, 2007
|
|
2OLW
 
 | | Crystal Structure of E. coli pseudouridine synthase RluE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETIC ACID, Ribosomal large subunit pseudouridine synthase E, ... | | Authors: | Pan, H, Ho, J.D, Stroud, R.M, Finer-Moore, J. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of E. coli rRNA Pseudouridine Synthase RluE.
J.Mol.Biol., 367, 2007
|
|
2P2R
 
 | | Crystal structure of the third KH domain of human Poly(C)-Binding Protein-2 in complex with C-rich strand of human telomeric DNA | | Descriptor: | 6-AMINOPYRIMIDIN-2(1H)-ONE, C-rich strand of human telomeric DNA, Poly(rC)-binding protein 2 | | Authors: | James, T.L, Stroud, R.M, Du, Z, Fenn, S, Tjhen, R, Lee, J.K. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-12 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the third KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.6 A resolution.
Nucleic Acids Res., 35, 2007
|
|
2Q9C
 
 | | Structure of FTSY:GMPPNP with MGCL Complex | | Descriptor: | Cell division protein ftsY, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, SULFATE ION | | Authors: | Reyes, C.L, Stroud, R.M. | | Deposit date: | 2007-06-12 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Structures of the Signal Recognition Particle Receptor Reveal Targeting Cycle Intermediates.
Plos One, 2, 2007
|
|
2Q9B
 
 | | Structure of FTSY:GMPPNP Complex | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein ftsY, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Reyes, C.L, Stroud, R.M. | | Deposit date: | 2007-06-12 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Structures of the Signal Recognition Particle Receptor Reveal Targeting Cycle Intermediates.
Plos One, 2, 2007
|
|
2Q9A
 
 | | Structure of Apo FTSY | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein ftsY, SULFATE ION | | Authors: | Reyes, C.L, Stroud, R.M. | | Deposit date: | 2007-06-12 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | X-ray Structures of the Signal Recognition Particle Receptor Reveal Targeting Cycle Intermediates.
Plos One, 2, 2007
|
|
2SAM
 
 | | STRUCTURE OF THE PROTEASE FROM SIMIAN IMMUNODEFICIENCY VIRUS: COMPLEX WITH AN IRREVERSIBLE NON-PEPTIDE INHIBITOR | | Descriptor: | 3-(4-NITRO-PHENOXY)-PROPAN-1-OL, SIV PROTEASE | | Authors: | Rose, R.B, Rose, J.R, Salto, R, Craik, C.S, Stroud, R.M. | | Deposit date: | 1994-07-08 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the protease from simian immunodeficiency virus: complex with an irreversible nonpeptide inhibitor.
Biochemistry, 32, 1993
|
|
7KP4
 
 | |
7JXF
 
 | | E. coli TSase complex with a bi-substrate reaction intermediate diastereomer analog | | Descriptor: | (2S)-2-({4-[({(6S)-2,4-diamino-5-[(1-{(2R,4S,5R)-4-hydroxy-5-[(phosphonooxy)methyl]tetrahydrofuran-2-yl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)methyl]-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl}methyl)amino]benzoyl}amino)pentanedioic acid (non-preferred name), 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE, N-[4-({[(6S)-2,4-diamino-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl]methyl}amino)benzene-1-carbonyl]-L-glutamic acid, ... | | Authors: | Finer-Moore, J, Kholodar, S.A, Stroud, R.M, Kohen, A, Moliner, V, Swiderek, K. | | Deposit date: | 2020-08-27 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caught in Action: X-ray Structure of Thymidylate Synthase with Noncovalent Intermediate Analog.
Biochemistry, 60, 2021
|
|
7JX1
 
 | | E. coli TSase complex with a bi-substrate reaction intermediate analog | | Descriptor: | (2S)-2-({4-[({(6R)-2,4-diamino-5-[(1-{(2R,4S,5R)-4-hydroxy-5-[(phosphonooxy)methyl]tetrahydrofuran-2-yl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)methyl]-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl}methyl)amino]benzoyl}amino)pentanedioic acid (non-preferred name), N-(4-{[(2,4-diamino-7,8-dihydropyrido[3,2-d]pyrimidin-6-yl)methyl]amino}benzene-1-carbonyl)-L-glutamic acid, PHOSPHATE ION, ... | | Authors: | Finer-Moore, J, Kholodar, S.A, Stroud, R.M, Kohen, A. | | Deposit date: | 2020-08-26 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Caught in Action: X-ray Structure of Thymidylate Synthase with Noncovalent Intermediate Analog.
Biochemistry, 60, 2021
|
|
7KTX
 
 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to a Fab in DDM detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-24 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7KRA
 
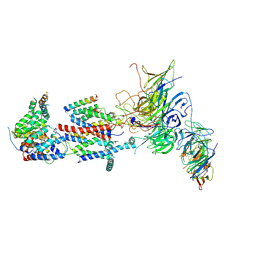 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to Fab-DH4 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-19 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
1SQG
 
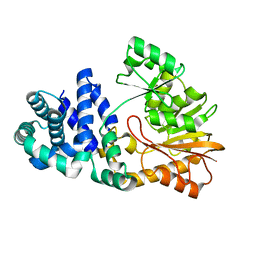 | | The crystal structure of the E. coli Fmu apoenzyme at 1.65 A resolution | | Descriptor: | SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
1TDU
 
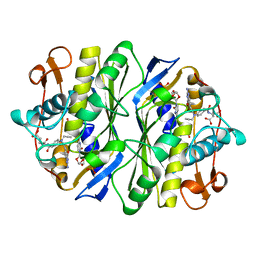 | | E. COLI THYMIDYLATE SYNTHASE IN COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE (DURD) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE, PHOSPHATE ION, ... | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-25 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
