3R2F
 
 | | Crystal structure of beta-site app-cleaving enzyme 1 (BACE-WT) complex with BMS-693391 AKA (2S)-2-((3R)-3-acetamido-3-isobutyl-2-oxo-1-pyrrolidinyl)-N-((1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-2-((2R,4R)-4-propoxy-2-pyrrolidinyl)ethyl)-4-phenylbutanamide | | Descriptor: | (2S)-2-[(3R)-3-(acetylamino)-3-(2-methylpropyl)-2-oxopyrrolidin-1-yl]-N-{(1R,2S)-3-(3,5-difluorophenyl)-1-hydroxy-1-[(2R,4R)-4-propoxypyrrolidin-2-yl]propan-2-yl}-4-phenylbutanamide, Beta-secretase 1 | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2011-03-14 | | Release date: | 2011-08-31 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Monosubstituted {gamma}-lactam and conformationally constrained 1,3-diaminopropan-2-ol transition-state isostere inhibitors of {beta}-secretase (BACE).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1I9D
 
 | | ARSENATE REDUCTASE FROM E. COLI | | Descriptor: | ARSENATE REDUCTASE, CESIUM ION, SULFATE ION, ... | | Authors: | Martin, P, Edwards, B.F. | | Deposit date: | 2001-03-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme.
Structure, 9, 2001
|
|
8K4B
 
 | | Cryo-EM structure of nucleotide-bound ComA with ZinC ion | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Competence factor transporting ATP-binding protein/permease ComA, ZINC ION | | Authors: | Yu, L, Xin, X, Min, L, Feng, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-11 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of peptide secretion for Quorum sensing by ComA
Nat Commun, 14, 2023
|
|
8K7A
 
 | |
7Y6F
 
 | | Cryo-EM structure of Apo form of ScBfr | | Descriptor: | Bacterioferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Bacterioferritin nanocage structures uncover the biomineralization process in ferritins.
Pnas Nexus, 2, 2023
|
|
7Y6P
 
 | | Cryo-EM structure if bacterioferritin holoform | | Descriptor: | Bacterioferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Bacterioferritin nanocage structures uncover the biomineralization process in ferritins.
Pnas Nexus, 2, 2023
|
|
7Y6G
 
 | |
1J9B
 
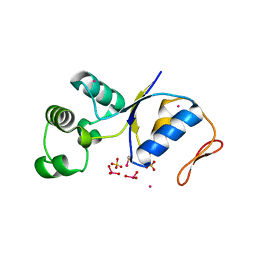 | | ARSENATE REDUCTASE+0.4M ARSENITE FROM E. COLI | | Descriptor: | ARSENATE REDUCTASE, CESIUM ION, SULFATE ION, ... | | Authors: | Martin, P, Edwards, B.F. | | Deposit date: | 2001-05-24 | | Release date: | 2001-12-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme.
Structure, 9, 2001
|
|
8H02
 
 | |
8HF4
 
 | |
8HF5
 
 | |
8HF6
 
 | | Cryo-EM structure of nucleotide-bound ComA E647Q mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Competence factor transporting ATP-binding protein/permease ComA | | Authors: | Yu, L, Xin, X, Min, L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of peptide secretion for Quorum sensing by ComA.
Nat Commun, 14, 2023
|
|
8HF7
 
 | |
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | Descriptor: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | Deposit date: | 2015-10-07 | | Release date: | 2016-03-16 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
1S3D
 
 | | ARSENATE REDUCTASE R60A MUTANT FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION | | Authors: | Demel, S, Edwards, B.F. | | Deposit date: | 2004-01-13 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
1S3C
 
 | | ARSENATE REDUCTASE C12S MUTANT FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION | | Authors: | DeMel, S, Edwards, B.F. | | Deposit date: | 2004-01-13 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
1SD8
 
 | | ARSENATE REDUCTASE R60K MUTANT FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION | | Authors: | DeMel, S, Edwards, B.F. | | Deposit date: | 2004-02-13 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
1SD9
 
 | | ARSENATE REDUCTASE C12S MUTANT +0.4M ARSENATE FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION | | Authors: | DeMel, S, Edwards, B.F. | | Deposit date: | 2004-02-13 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
1SK1
 
 | | ARSENATE REDUCTASE R60K MUTANT +0.4M ARSENATE FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION | | Authors: | DeMel, S, Edwards, B.F. | | Deposit date: | 2004-03-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
6IWI
 
 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | MAGNESIUM ION, N-[3-(4,5-diethyl-6-oxo-1,6-dihydropyrimidin-2-yl)-4-propoxyphenyl]-2-(4-methylpiperazin-1-yl)acetamide, ZINC ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Pharmacokinetics-Driven Optimization of 4(3 H)-Pyrimidinones as Phosphodiesterase Type 5 Inhibitors Leading to TPN171, a Clinical Candidate for the Treatment of Pulmonary Arterial Hypertension.
J.Med.Chem., 62, 2019
|
|
7TXC
 
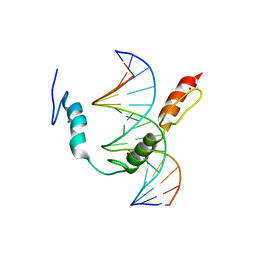 | | HIC2 zinc finger domain in complex with the DNA binding motif-2 of the BCL11A enhancer | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*TP*GP*GP*CP*AP*TP*TP*AP*TP*CP*T)-3'), DNA (5'-D(*AP*GP*AP*TP*AP*AP*TP*GP*CP*CP*AP*AP*CP*AP*GP*T)-3'), Hypermethylated in cancer 2 protein, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-01 | | Last modified: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | HIC2 controls developmental hemoglobin switching by repressing BCL11A transcription.
Nat.Genet., 54, 2022
|
|
7V3F
 
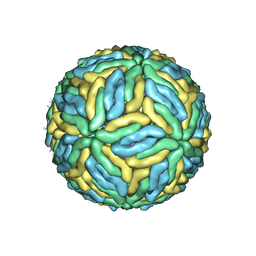 | | DENV2_NGC_Fab_C10 28degree (1Fab:3E) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Shu, B, Zhang, S, Victor, A.K, Ng, T.S, Lok, S.M. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Human antibody C10 neutralizes by diminishing Zika but enhancing dengue virus dynamics.
Cell, 184, 2021
|
|
7V3J
 
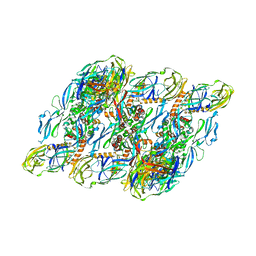 | | DENV2:F(ab')2-local | | Descriptor: | Envelope protein E, Fab_C10_heavy_chain, Fab_C10_light_chain, ... | | Authors: | Shu, B, Zhang, S, Victor, A.K, Ng, T.S, Lok, S.M. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Human antibody C10 neutralizes by diminishing Zika but enhancing dengue virus dynamics.
Cell, 184, 2021
|
|
7V3I
 
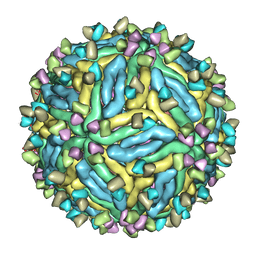 | | DENV2_NGC_Fab_C10 4degrees (3Fab:3E) | | Descriptor: | Envelope protein E, Fab_C10_heavy_chain, Fab_C10_light_chain, ... | | Authors: | Shu, B, Zhang, S, Victor, A.K, Ng, T.S, Lok, S.M. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Human antibody C10 neutralizes by diminishing Zika but enhancing dengue virus dynamics.
Cell, 184, 2021
|
|
7V3G
 
 | | DENV2_NGC_Fab_C10 28degrees (2Fab:3E) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Shu, B, Zhang, S, Victor, A.K, Ng, T.S, Lok, S.M. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Human antibody C10 neutralizes by diminishing Zika but enhancing dengue virus dynamics.
Cell, 184, 2021
|
|
