5UEW
 
 | | BRD2 Bromodomain2 with A-1360579 | | Descriptor: | Bromodomain-containing protein 2, N-[3-(4-methoxy-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-4-phenoxyphenyl]methanesulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEY
 
 | | BRD4_BD2_A-1412838 | | Descriptor: | 5-[2-(2,4-difluorophenoxy)-5-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}phenyl]-4-ethoxy-1-methylpyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEX
 
 | | BRD4_BD2_A-1497627 | | Descriptor: | 17-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}-11,13-difluoro-2-methyl-6,7,8,9-tetrahydrodibenzo[4,5:7,8][1,6]dioxacyclododecino[3,2-c]pyridin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEU
 
 | | BRD4_BD2_A-1107604 | | Descriptor: | Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UF0
 
 | | BRD4_BD2-A-35165 | | Descriptor: | 2-methyl-5-(methylamino)-6-phenylpyridazin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEZ
 
 | | BRD4_BD2_A-1342843 | | Descriptor: | 5-methoxy-2-methyl-6-(2-phenoxyphenyl)pyridazin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5WQT
 
 | | Structure of a protein involved in pyroptosis | | Descriptor: | CITRIC ACID, GLYCEROL, Gasdermin-D | | Authors: | Kuang, S, Li, J. | | Deposit date: | 2016-11-28 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4KSA
 
 | | Crystal Structure of Malonyl-CoA decarboxylase from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR127 | | Descriptor: | MAGNESIUM ION, Malonyl-CoA decarboxylase | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Patel, D.J, Ciccosanti, C, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of malonyl-coenzyme a decarboxylase provide insights into its catalytic mechanism and disease-causing mutations.
Structure, 21, 2013
|
|
7XDY
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDW
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDV
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDX
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7E39
 
 | | SARS-CoV-2 spike in complex with the Ab4 neutralizing antibody (State 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Ab4, Light Chain of Ab4, ... | | Authors: | Liu, C. | | Deposit date: | 2021-02-08 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Three epitope-distinct human antibodies from RenMab mice neutralize SARS-CoV-2 and cooperatively minimize the escape of mutants.
Cell Discov, 7, 2021
|
|
7E3B
 
 | |
7E3C
 
 | |
7DV4
 
 | | Crystal structure of anti-CTLA-4 VH domain in complex with human CTLA-4 | | Descriptor: | 1,2-ETHANEDIOL, 4003-1(VH), Cytotoxic T-lymphocyte protein 4 | | Authors: | Li, H, Gan, X, He, Y. | | Deposit date: | 2021-01-12 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | An anti-CTLA-4 heavy chain-only antibody with enhanced T reg depletion shows excellent preclinical efficacy and safety profile.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVM
 
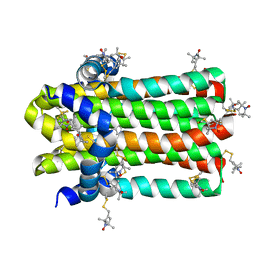 | | DgkA structure in E.coli lipid bilayer | | Descriptor: | Diacylglycerol kinase | | Authors: | Li, J, Yang, J. | | Deposit date: | 2021-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2023-09-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of membrane diacylglycerol kinase in lipid bilayers.
Commun Biol, 4, 2021
|
|
5WYB
 
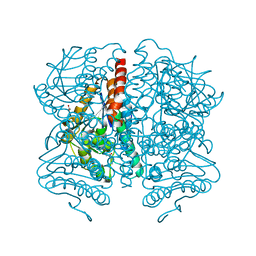 | | Structure of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
5WYD
 
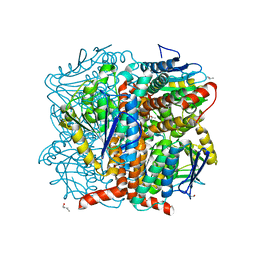 | | Structural of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
7CO5
 
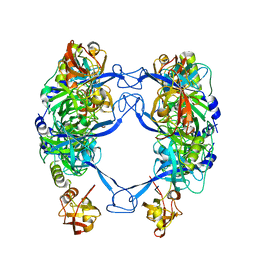 | | HtrA-type protease AlgW with decapeptide | | Descriptor: | AlgW protein, IMIDAZOLE, decapeptide SVRDELRWVF | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO3
 
 | | HtrA-type protease AlgWS227A with tripeptide | | Descriptor: | AlgW protein, TRP-VAL-PHE | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO2
 
 | | HtrA-type protease AlgW with tripeptide | | Descriptor: | AlgW protein, IMIDAZOLE, TRP-VAL-PHE | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO7
 
 | | HtrA-type protease AlgWS227A with decapeptide | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, AlgW protein, decapeptide SVRDELRWVF | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-04 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CZP
 
 | | S protein of SARS-CoV-2 in complex bound with P2B-1A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c181_light_IGLV2-14_IGLJ3,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZQ
 
 | | S protein of SARS-CoV-2 in complex bound with P2B-1A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c642_heavy_IGHV3-53_IGHD1-26_IGHJ6,Chain H of P2B-1A10,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
