6XSP
 
 | |
6XSQ
 
 | |
6XT3
 
 | |
6WHD
 
 | | Crystal structure of E.coli DsbA in complex with diaryl ether analogue 2 | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, [4-(4-cyano-3-methylphenoxy)phenyl]acetic acid | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiLX).
J.Med.Chem., 63, 2020
|
|
1PT4
 
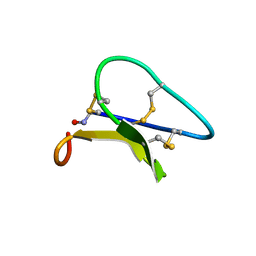 | | Solution structure of the Moebius cyclotide kalata B2 | | Descriptor: | kalata B2 | | Authors: | Jennings, C.V, Anderson, M.A, Daly, N.L, Rosengren, K.J, Craik, D.J. | | Deposit date: | 2003-06-23 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Isolation, Solution Structure, and Insecticidal Activity of Kalata B2, a Circular Protein with a Twist: Do Mobius Strips Exist in Nature?(,)
Biochemistry, 44, 2005
|
|
2MIM
 
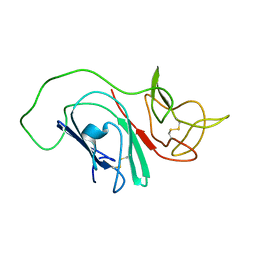 | |
4ZL7
 
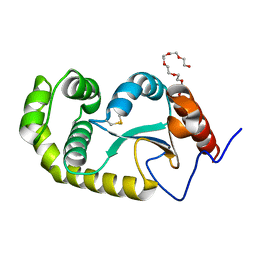 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal I | | Descriptor: | HEXAETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZL9
 
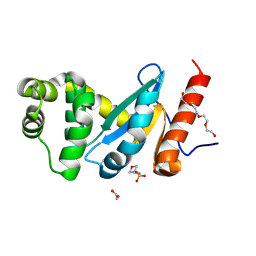 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal III | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZL8
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | McMahoh, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
2MHC
 
 | | NMR structure of the catalytic domain of the large serine resolvase TnpX | | Descriptor: | TnpX | | Authors: | Headey, S.J, Sivakumaran, A, Adams, V, Rodgers, A.J.W, Rood, J.I, Scanlon, M.J, Wilce, M.C.J. | | Deposit date: | 2013-11-20 | | Release date: | 2014-11-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and DNA Binding of the Catalytic of the Large Serine Resolvase Tnpx
To be Published
|
|
4DVC
 
 | | Structural and functional studies of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera toxin production | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Walden, P.M, Martin, J.L. | | Deposit date: | 2012-02-23 | | Release date: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A resolution crystal structure of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera-toxin production
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3WNG
 
 | |
3WNE
 
 | |
3WNF
 
 | |
3WNH
 
 | |
7KRI
 
 | | FR6-bound SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | 1,3-dimethyl-1H-pyrrolo[3,4-d]pyrimidine-2,4(3H,6H)-dione, MALONATE ION, Non-structural protein 9, ... | | Authors: | Littler, D.R, Gully, B.S, Rossjohn, J. | | Deposit date: | 2020-11-20 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding of a pyrimidine RNA base-mimic to SARS-CoV-2 nonstructural protein 9.
J.Biol.Chem., 297, 2021
|
|
7L23
 
 | |
1F5Y
 
 | | NMR STRUCTURE OF A CONCATEMER OF THE FIRST AND SECOND LIGAND-BINDING MODULES OF THE HUMAN LDL RECEPTOR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Kurniawan, N.D, Atkins, A.R, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2000-06-18 | | Release date: | 2000-08-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a concatemer of the first and second ligand-binding modules of the human low-density lipoprotein receptor.
Protein Sci., 9, 2000
|
|
