8BQ3
 
 | |
1AYE
 
 | | HUMAN PROCARBOXYPEPTIDASE A2 | | Descriptor: | PROCARBOXYPEPTIDASE A2, ZINC ION | | Authors: | Garcia-Saez, I, Reverte, D, Vendrell, J, Aviles, F.X, Coll, M. | | Deposit date: | 1997-11-03 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of human procarboxypeptidase A2. Deciphering the basis of the inhibition, activation and intrinsic activity of the zymogen.
EMBO J., 16, 1997
|
|
7VIM
 
 | |
4BWR
 
 | | Crystal structure of c5321: a protective antigen present in uropathogenic Escherichia coli strains displaying an SLR fold | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Urosev, D, Ferrer-Navarro, M, Pastorello, I, Cartocci, E, Costenaro, L, Zhulenkovs, D, Marechal, J.-D, Leonchiks, A, Reverter, D, Serino, L, Soriani, M, Daura, X. | | Deposit date: | 2013-07-04 | | Release date: | 2013-07-24 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of C5321: A Protective Antigen Present in Uropathogenic Escherichia Coli Strains Displaying an Slr Fold.
Bmc Struct.Biol., 13, 2013
|
|
8PM8
 
 | | V30M Transthyretin structure in complex with Tolcalpone | | Descriptor: | Tolcapone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
8PMA
 
 | | Crystal structure of human V30M transthyretin in complex with PITB (Pharmacokinetically Improved TTR Binder) | | Descriptor: | (3-fluoranyl-5-oxidanyl-phenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
8PM9
 
 | | Crystal structure of human wild type transthyretin in complex with PITB (Pharmacokinetically Improved TTR Binder) | | Descriptor: | (3-fluoranyl-5-oxidanyl-phenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
8PMO
 
 | | Crystal structure of human V122I transthyretin in complex with PITB (Pharmacokinetically Improved TTR Binder) | | Descriptor: | (3-fluoranyl-5-oxidanyl-phenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
5OM9
 
 | | Crystal structure of the human CARBOXYPEPTIDASE A1 in complex with a thiirane mechanism-based inhibitor | | Descriptor: | (2~{R})-4-methyl-2-[(1~{S})-1-sulfanylethyl]pentanoic acid, Carboxypeptidase A1, ZINC ION | | Authors: | Gallego, P, Granados, C, Fernandez, D, Pallares, I, Covaleda, G, Aviles, F.X, Vendrell, J, Reverter, D. | | Deposit date: | 2017-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Mechanism-Based Inactivators for Human Pancreatic Carboxypeptidase A from a Focused Synthetic Library.
ACS Med Chem Lett, 8, 2017
|
|
5AEK
 
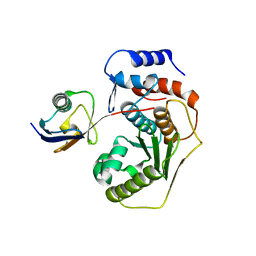 | | Crystal structure of the human SENP2 C548S in complex with the human SUMO1 K48M F66W | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 2, SMALL UBIQUITIN-RELATED MODIFIER 1 | | Authors: | Gallego, P, Grana-Montes, R, Espargaro, A, Castillo, V, Torrent, J, Lange, R, Papaleo, E, Lindorff-Larsend, K, Ventura, S, Reverter, D. | | Deposit date: | 2014-12-23 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stepping Back and Forward on Sumo Folding Evolution
To be Published
|
|
5AQ0
 
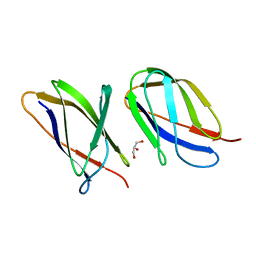 | | The structure of the Transthyretin-like domain of the first catalytic domain of the HUMAN Carboxypeptidase D | | Descriptor: | CARBOXYPEPTIDASE D, GLYCEROL | | Authors: | Gallego, P, Garcia-Pardo, J, Lorenzo, J, Aviles, F.X, Ventura, S, Reverter, D. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The Structure of the Ttldomain of the Human Carboxypeptidase D
To be Published
|
|
5LCN
 
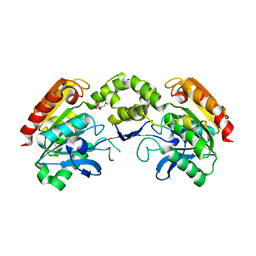 | |
6FWW
 
 | | GFP/KKK. A redesigned GFP with improved solubility | | Descriptor: | Green fluorescent protein | | Authors: | Varejao, N, Lascorz, J, Gil-Garcia, M, Diaz-Caballero, M, Navarro, S, Ventura, S, Reverter, D. | | Deposit date: | 2018-03-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Combining Structural Aggregation Propensity and Stability Predictions To Redesign Protein Solubility.
Mol. Pharm., 15, 2018
|
|
6FXU
 
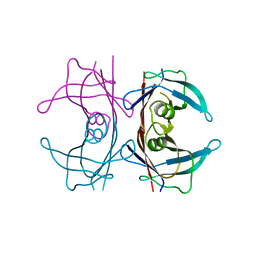 | |
6FZL
 
 | |
7AMN
 
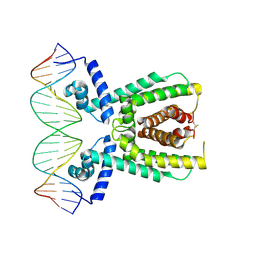 | | Structure of LuxR with DNA (repression) | | Descriptor: | DNA (5'-D(P*TP*AP*TP*TP*GP*AP*TP*AP*AP*AP*AP*TP*TP*AP*TP*CP*AP*AP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*TP*GP*AP*TP*AP*AP*TP*TP*TP*TP*AP*TP*CP*AP*AP*TP*A)-3'), HTH-type transcriptional regulator LuxR | | Authors: | Liu, B, Reverter, D. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
7AMT
 
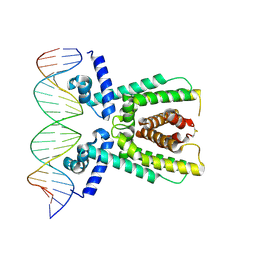 | | Structure of LuxR with DNA (activation) | | Descriptor: | DNA (5'-D(P*AP*TP*AP*AP*TP*GP*AP*CP*AP*TP*TP*AP*CP*TP*GP*TP*AP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*TP*AP*CP*AP*GP*TP*AP*AP*TP*GP*TP*CP*AP*TP*TP*AP*T)-3'), HTH-type transcriptional regulator LuxR | | Authors: | Liu, B, Reverter, D. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
5MRV
 
 | | Crystal structure of human carboxypeptidase O in complex with NvCI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase O, Metallocarboxypeptidase inhibitor, ... | | Authors: | Garcia-Pardo, J, Garcia-Guerrero, M.C, Fernandez-Alvarez, R, Lyons, P, Aviles, F.X, Lorenzo, J, Reverter, D. | | Deposit date: | 2016-12-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Crystal structure and mechanism of human carboxypeptidase O: Insights into its specific activity for acidic residues.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4E4J
 
 | | Crystal structure of arginine deiminase from Mycoplasma penetrans | | Descriptor: | Arginine deiminase, CHLORIDE ION | | Authors: | Benach, J, Gallego, P, Planell, R, Querol, E, Perez Pons, J.A, Reverter, D. | | Deposit date: | 2012-03-13 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma penetrans.
Plos One, 7, 2012
|
|
3EAY
 
 | | Crystal structure of the human SENP7 catalytic domain | | Descriptor: | SULFATE ION, Sentrin-specific protease 7 | | Authors: | Lima, C.D, Reverter, D. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Human SENP7 Catalytic Domain and Poly-SUMO Deconjugation Activities for SENP6 and SENP7.
J.Biol.Chem., 283, 2008
|
|
1JQG
 
 | | Crystal Structure of the Carboxypeptidase A from Helicoverpa Armigera | | Descriptor: | ZINC ION, carboxypeptidase A | | Authors: | Estebanez-Perpina, E, Bayes, A, Vendrell, J, Jongsma, M.A, Bown, D.P, Gatehouse, J.A, Huber, R, Bode, W, Aviles, F.X, Reverter, D. | | Deposit date: | 2001-08-07 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a novel mid-gut procarboxypeptidase from the cotton pest Helicoverpa armigera.
J.Mol.Biol., 313, 2001
|
|
5FW6
 
 | | Structure of human transthyretin mutant A108V | | Descriptor: | TRANSTHYRETIN | | Authors: | Gallego, P, Varejao, N, Santanna, R, Saraiva, M.J, Ventura, S, Reverter, D. | | Deposit date: | 2016-02-12 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Cavity filling mutations at the thyroxine-binding site dramatically increase transthyretin stability and prevent its aggregation.
Sci Rep, 7, 2017
|
|
5FO2
 
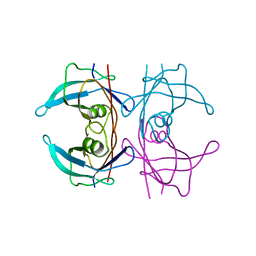 | | Structure of human transthyretin mutant A108I | | Descriptor: | TRANSTHYRETIN | | Authors: | Varejao, N, Santanna, R, Saraiva, M.J, Gallego, P, Ventura, S, Reverter, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Cavity filling mutations at the thyroxine-binding site dramatically increase transthyretin stability and prevent its aggregation.
Sci Rep, 7, 2017
|
|
5G5C
 
 | |
5G59
 
 | |
