1VG1
 
 | | GDP-Bound Rab7 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-7 | | Authors: | Rak, A, Pylypenko, O, Niculae, A, Pyatkov, K, Goody, R.S, Alexandrov, K. | | Deposit date: | 2004-04-22 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease
Cell(Cambridge,Mass.), 117, 2004
|
|
1VG0
 
 | | The crystal structures of the REP-1 protein in complex with monoprenylated Rab7 protein | | Descriptor: | CHLORIDE ION, GERAN-8-YL GERAN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rak, A, Pylypenko, O, Niculae, A, Pyatkov, K, Goody, R.S, Alexandrov, K. | | Deposit date: | 2004-04-22 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease
Cell(Cambridge,Mass.), 117, 2004
|
|
1VG9
 
 | | The crystal structures of the REP-1 protein in complex with C-terminally truncated Rab7 protein | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rak, A, Pylypenko, O, Niculae, A, Pyatkov, K, Goody, R.S, Alexandrov, K. | | Deposit date: | 2004-04-23 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease
Cell(Cambridge,Mass.), 117, 2004
|
|
1N3G
 
 | |
1UKV
 
 | | Structure of RabGDP-dissociation inhibitor in complex with prenylated YPT1 GTPase | | Descriptor: | GERAN-8-YL GERAN, GTP-binding protein YPT1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rak, A, Pylypenko, O, Durek, T, Watzke, A, Kushnir, S, Brunsveld, L, Waldmann, H, Goody, R.S, Alexandrov, K. | | Deposit date: | 2003-09-01 | | Release date: | 2004-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase
Science, 302, 2003
|
|
1VG8
 
 | | GPPNHP-Bound Rab7 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-7 | | Authors: | Rak, A, Pylypenko, O, Niculae, A, Pyatkov, K, Goody, R.S, Alexandrov, K. | | Deposit date: | 2004-04-23 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease
Cell(Cambridge,Mass.), 117, 2004
|
|
1FKM
 
 | | CRYSTAL STRUCTURE OF THE YPT/RAB-GAP DOMAIN OF GYP1P | | Descriptor: | PROTEIN (GYP1P) | | Authors: | Rak, A, Fedorov, R, Alexandrov, K, Albert, S, Goody, R.S, Gallwitz, D, Scheidig, A.J. | | Deposit date: | 2000-08-09 | | Release date: | 2001-02-09 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins.
EMBO J., 19, 2000
|
|
1CTN
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL CHITINASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CHITINASE A | | Authors: | Perrakis, A, Tews, I, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bacterial chitinase at 2.3 A resolution.
Structure, 2, 1994
|
|
4MB7
 
 | |
4NRV
 
 | | Crystal Structure of non-edited human NEIL1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endonuclease 8-like 1 | | Authors: | Prakash, A, Doublie, S. | | Deposit date: | 2013-11-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: Activity, structure, and the effect of editing.
Dna Repair, 14, 2014
|
|
4NRW
 
 | | MvNei1-G86D | | Descriptor: | 5'-D(*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*C)-3', 5'-D(*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*G)-3', formamidopyrimidine-DNA glycosylase | | Authors: | Prakash, A, Doublie, S. | | Deposit date: | 2013-11-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: Activity, structure, and the effect of editing.
Dna Repair, 14, 2014
|
|
3ATU
 
 | | Crystal structure of human Hsp70 NBD in the ADP- and Mg ion-bound state | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1A/1B, ... | | Authors: | Arakawa, A, Handa, N, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-13 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical and structural studies on the high affinity of Hsp70 for ADP.
Protein Sci., 20, 2011
|
|
3ATV
 
 | | Crystal structure of human Hsp70 NBD in the ADP-bound and Mg ion-free state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 1A/1B, ... | | Authors: | Arakawa, A, Handa, N, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Biochemical and structural studies on the high affinity of Hsp70 for ADP.
Protein Sci., 20, 2011
|
|
3AY9
 
 | | Crystal structure of human Hsp70 NBD in the ADP-, Mg ion-, and K ion-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Arakawa, A, Handa, N, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-05-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural studies on the high affinity of Hsp70 for ADP.
Protein Sci., 20, 2011
|
|
3A8Y
 
 | | Crystal structure of the complex between the BAG5 BD5 and Hsp70 NBD | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG family molecular chaperone regulator 5, Heat shock 70 kDa protein 1 | | Authors: | Arakawa, A, Handa, N, Ohsawa, N, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-13 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
8BW0
 
 | | Structure of CEACAM5 A3-B3 domain in Complex with Tusamitamab Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 5, ... | | Authors: | Kumar, A, Bertrand, T, Rapisarda, C, Rak, A. | | Deposit date: | 2022-12-06 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insights into epitope-paratope interactions of monoclonal antibody targeting CEACAM5-expressing tumors
Res Sq, 2023
|
|
6QCG
 
 | |
5D75
 
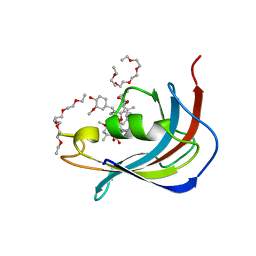 | | Crystal structure of Human FKBD25 in complex with FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), Peptidyl-prolyl cis-trans isomerase FKBP3 | | Authors: | Rajan, S, Prakash, A, Yoon, H.S. | | Deposit date: | 2015-08-13 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the FK506 binding domain of human FKBP25 in complex with FK506.
Protein Sci., 25, 2016
|
|
7BFZ
 
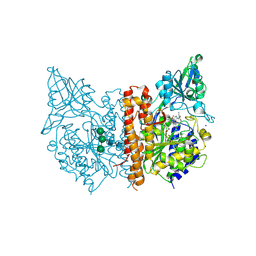 | | X-ray structure of human prostate-specific membrane antigen(PSMA) in complex with a inhibitor Glu-490 | | Descriptor: | (((S)-1-carboxy-5-((E)-2-cyano-3-(5-(1-(3-methoxy-3-oxopropyl)-1,2,3,4-tetrahydroquinolin-6-yl)thiophen-2-yl)acrylamido)pentyl)carbamoyl)-L-glutamic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rakhimbekova, A, Motlova, L, Barinka, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A prostate-specific membrane antigen activated molecular rotor for real-time fluorescence imaging.
Nat Commun, 12, 2021
|
|
8ARF
 
 | |
2MPH
 
 | |
1LTX
 
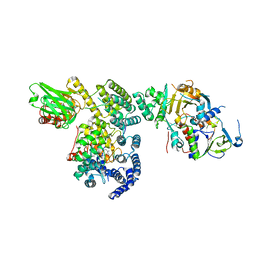 | | Structure of Rab Escort Protein-1 in complex with Rab geranylgeranyl transferase and isoprenoid | | Descriptor: | AAAA, CHLORIDE ION, FARNESYL, ... | | Authors: | Pylypenko, O, Rak, A, Reents, R, Niculae, A, Thoma, N.H, Waldmann, H, Schlichting, I, Goody, R.S, Alexandrov, K. | | Deposit date: | 2002-05-21 | | Release date: | 2003-05-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Rab Escort Protein-1 in Complex with Rab Geranylgeranyltransferase
Mol.Cell, 11, 2003
|
|
8TYL
 
 | |
8TYO
 
 | |
4UUQ
 
 | | Crystal structure of human mono-glyceride lipase in complex with SAR127303 | | Descriptor: | 4-({[(4-chlorophenyl)sulfonyl]amino}methyl)piperidine-1-carboxylic acid, MONOGLYCERIDE LIPASE | | Authors: | Griebel, G, Pichat, P, Beeske, S, Leroy, T, Redon, N, Francon, D, Bert, L, Even, L, Lopez-Grancha, M, Tolstykh, T, Sun, F, Yu, Q, Brittain, S, Arlt, H, He, T, Zhang, B, Wiederschain, D, Bertrand, T, Houtman, J, Rak, A, Vallee, F, Michot, N, Auge, F, Menet, V, Bergis, O.E, George, P, Avenet, P, Mikol, V, Didier, M, Escoubet, J. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Selective Blockade of the Hydrolysis of the Endocannabinoid 2-Arachidonoylglycerol Impairs Learning and Memory Performance While Producing Antinociceptive Activity in Rodents.
Sci.Rep., 5, 2015
|
|
