2PK9
 
 | | Structure of the Pho85-Pho80 CDK-cyclin Complex of the Phosphate-responsive Signal Transduction Pathway | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cyclin-dependent protein kinase PHO85, PHO85 cyclin PHO80 | | Authors: | Huang, K, Ferrin-O'Connell, I, Zhang, W, Leonard, G.A, O'Shea, E.K, Quiocho, F.A. | | Deposit date: | 2007-04-17 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structure of the Pho85-Pho80 CDK-Cyclin Complex of the Phosphate-Responsive Signal Transduction Pathway
Mol.Cell, 28, 2007
|
|
2PMI
 
 | | Structure of the Pho85-Pho80 CDK-cyclin Complex of the Phosphate-responsive Signal Transduction Pathway with Bound ATP-gamma-S | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cyclin-dependent protein kinase PHO85, PHO85 cyclin PHO80, ... | | Authors: | Huang, K, Ferrin-O'Connell, I, Zhang, W, Leonard, G.A, O'Shea, E.K, Quiocho, F.A. | | Deposit date: | 2007-04-23 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Pho85-Pho80 CDK-Cyclin Complex of the Phosphate-Responsive Signal Transduction Pathway
Mol.Cell, 28, 2007
|
|
2R6G
 
 | | The Crystal Structure of the E. coli Maltose Transporter | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Maltose transport system permease protein malF, Maltose transport system permease protein malG, ... | | Authors: | Oldham, M.L, Khare, D, Quiocho, F.A, Davidson, A.L, Chen, J. | | Deposit date: | 2007-09-05 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a catalytic intermediate of the maltose transporter.
Nature, 450, 2007
|
|
1PBP
 
 | | FINE TUNING OF THE SPECIFICITY OF THE PERIPLASMIC PHOSPHATE TRANSPORT RECEPTOR: SITE-DIRECTED MUTAGENESIS, LIGAND BINDING, AND CRYSTALLOGRAPHIC STUDIES | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Wang, Z, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1994-07-20 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fine tuning the specificity of the periplasmic phosphate transport receptor. Site-directed mutagenesis, ligand binding, and crystallographic studies.
J.Biol.Chem., 269, 1994
|
|
1MDQ
 
 | |
1V39
 
 | |
1VP9
 
 | |
1VP3
 
 | |
1DMB
 
 | |
2GA9
 
 | | Crystal Structure of the Heterodimeric Vaccinia Virus Polyadenylate Polymerase with Bound ATP-gamma-S | | Descriptor: | CALCIUM ION, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Moure, C.M, Bowman, B.R, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2006-03-08 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the vaccinia virus polyadenylate polymerase heterodimer: insights into ATP selectivity and processivity.
Mol.Cell, 22, 2006
|
|
2GAF
 
 | | Crystal Structure of the Vaccinia Polyadenylate Polymerase Heterodimer (apo form) | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, Poly(A) polymerase catalytic subunit | | Authors: | Moure, C.M, Bowman, B.R, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2006-03-08 | | Release date: | 2006-05-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the vaccinia virus polyadenylate polymerase heterodimer: insights into ATP selectivity and processivity.
Mol.Cell, 22, 2006
|
|
3C0W
 
 | | I-SceI in complex with a bottom nicked DNA substrate | | Descriptor: | CALCIUM ION, DNA (5'-D(*DAP*DCP*DGP*DCP*DTP*DAP*DGP*DGP*DGP*DAP*DTP*DAP*DAP*DCP*DAP*DGP*DGP*DGP*DTP*DAP*DAP*DTP*DAP*DC)-3'), DNA (5'-D(*DGP*DTP*DAP*DTP*DTP*DAP*DCP*DCP*DCP*DTP*DGP*DTP*DTP*DAP*DT)-3'), ... | | Authors: | Moure, C.M, Gimble, F.S, Quiocho, F.A. | | Deposit date: | 2008-01-21 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of I-SceI complexed to nicked DNA substrates: snapshots of intermediates along the DNA cleavage reaction pathway.
Nucleic Acids Res., 36, 2008
|
|
3C0X
 
 | | I-SceI in complex with a top nicked DNA substrate | | Descriptor: | CALCIUM ION, DNA (5'-D(*DC*DAP*DCP*DGP*DCP*DTP*DAP*DGP*DGP*DGP*DAP*DTP*DAP*DA)-3'), DNA (5'-D(*DG*DGP*DTP*DAP*DTP*DTP*DAP*DCP*DCP*DCP*DTP*DGP*DTP*DTP*DAP*DTP*DCP*DCP*DCP*DTP*DAP*DGP*DCP*DGP*DT)-3'), ... | | Authors: | Moure, C.M, Gimble, F.S, Quiocho, F.A. | | Deposit date: | 2008-01-21 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of I-SceI complexed to nicked DNA substrates: snapshots of intermediates along the DNA cleavage reaction pathway.
Nucleic Acids Res., 36, 2008
|
|
1FKW
 
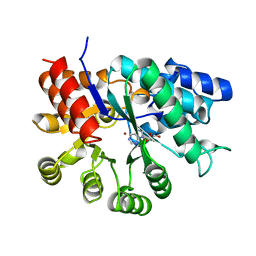 | | MURINE ADENOSINE DEAMINASE (D295E) | | Descriptor: | ADENOSINE DEAMINASE, PURINE RIBOSIDE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-02-29 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase.
Biochemistry, 35, 1996
|
|
1FKX
 
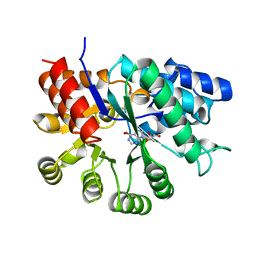 | | MURINE ADENOSINE DEAMINASE (D296A) | | Descriptor: | 6-HYDROXY-1,6-DIHYDRO PURINE NUCLEOSIDE, ADENOSINE DEAMINASE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-02-29 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase.
Biochemistry, 35, 1996
|
|
3MAG
 
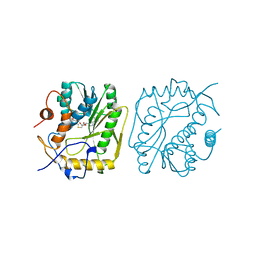 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M3ADE AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 6-AMINO-3-METHYLPURINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1998-07-12 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3MCT
 
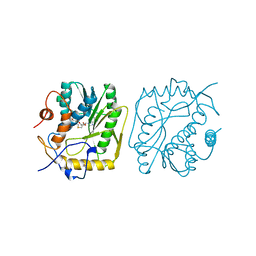 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M3CYT AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 3-METHYLCYTOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Gershon, P.D, Hodel, A.E, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1OIB
 
 | |
1PZ5
 
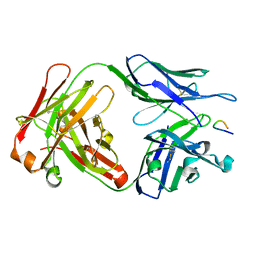 | | Structural basis of peptide-carbohydrate mimicry in an antibody combining site | | Descriptor: | Heavy chain of Fab (SYA/J6), Light chain of Fab (SYA/J6), Octapeptide (MDWNMHAA) | | Authors: | Vyas, N.K, Vyas, M.N, Chervenak, M.C, Bundle, D.R, Pinto, B.M, Quiocho, F.A. | | Deposit date: | 2003-07-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of peptide-carbohydrate mimicry in an antibody combining site.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1QUI
 
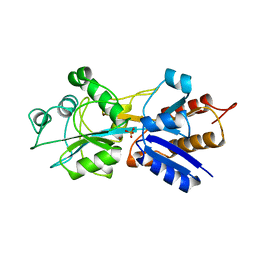 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY GLY COMPLEX WITH BROMINE AND PHOSPHATE | | Descriptor: | BROMIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUK
 
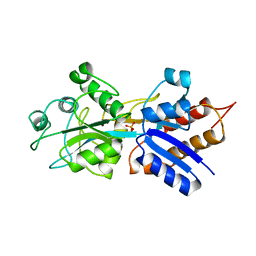 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY ASN COMPLEX WITH PHOSPHATE | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUJ
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY GLY COMPLEX WITH CHLORINE AND PHOSPHATE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1QUL
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY THR COMPLEX WITH CHLORINE AND PHOSPHATE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
1MDP
 
 | |
3RRK
 
 | | Crystal structure of the cytoplasmic N-terminal domain of subunit I, homolog of subunit a, of V-ATPase | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SULFATE ION, V-type ATPase 116 kDa subunit | | Authors: | Srinivasan, S, Vyas, N.K, Baker, M.L, Quiocho, F.A. | | Deposit date: | 2011-04-29 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the cytoplasmic N-terminal domain of subunit I, a homolog of subunit a, of V-ATPase.
J.Mol.Biol., 412, 2011
|
|
