8PD3
 
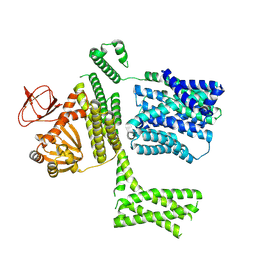 | | Ligand-free SpSLC9C1 in lipid nanodiscs, protomer state 2 | | Descriptor: | Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PDV
 
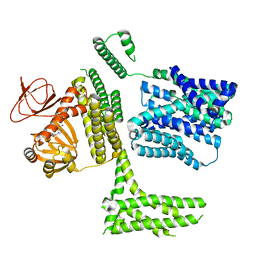 | | cGMP-bound SpSLC9C1 in lipid nanodiscs, protomer | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PCZ
 
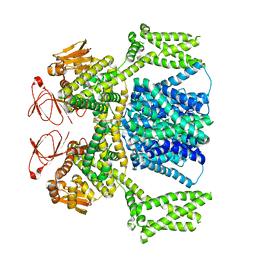 | | Ligand-free SpSLC9C1 in lipid nanodiscs, dimer | | Descriptor: | Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
7P7A
 
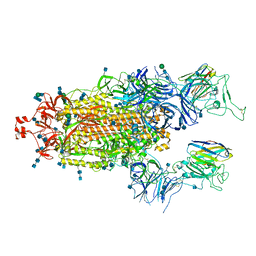 | | SARS-CoV-2 spike protein in complex with sybody#68 in a 2up/1flexible conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P77
 
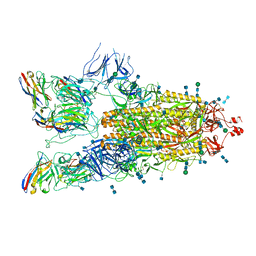 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 3up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P78
 
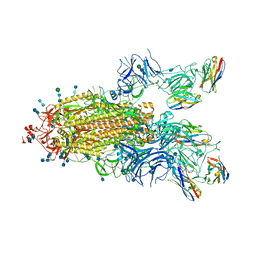 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 1up/1up-out/1down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P79
 
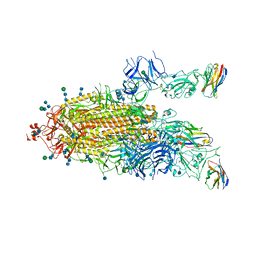 | | SARS-CoV-2 spike protein in complex with sybodyb#15 in a 1up/1up-out/1down conformation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P7B
 
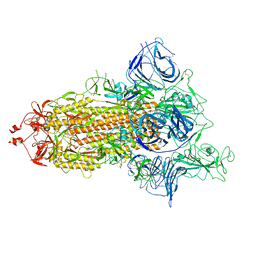 | | SARS-CoV-2 spike protein in complex with sybody no68 in a 1up/2down conformation | | Descriptor: | Spike glycoprotein | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7NNT
 
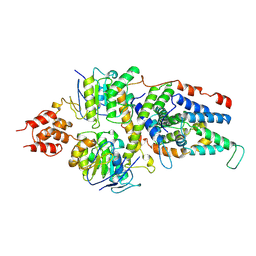 | | Cryo-EM structure of the folate-specific ECF transporter complex in DDM micelles | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2021-02-25 | | Release date: | 2021-08-18 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Insights into the bilayer-mediated toppling mechanism of a folate-specific ECF transporter by cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NNU
 
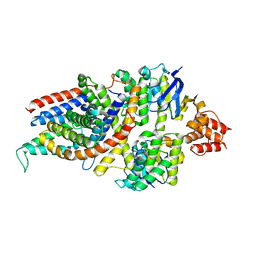 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs | | Descriptor: | Conserved hypothetical membrane protein, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2021-02-25 | | Release date: | 2021-08-18 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Insights into the bilayer-mediated toppling mechanism of a folate-specific ECF transporter by cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BCQ
 
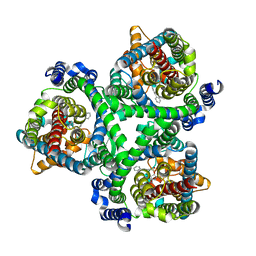 | | ASCT2 in the presence of the inhibitor Lc-BPE (position "up") in the outward-open conformation. | | Descriptor: | 4-(4-phenylphenyl)carbonyloxypyrrolidine-2-carboxylic acid, Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BCT
 
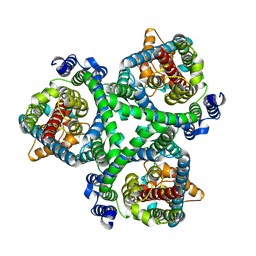 | | ASCT2 in the presence of the inhibitor ERA-21 in the outward-open conformation. | | Descriptor: | Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ZRI
 
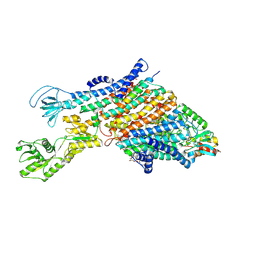 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRM
 
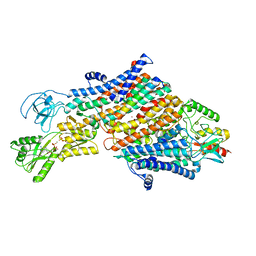 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRD
 
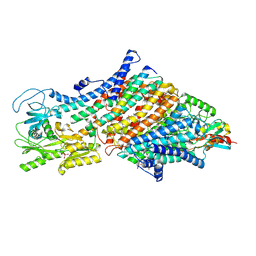 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, stabilised with the inhibitor orthovanadate | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRJ
 
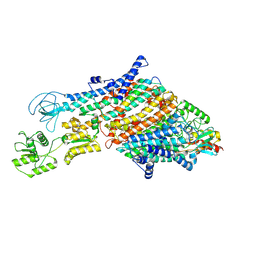 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRG
 
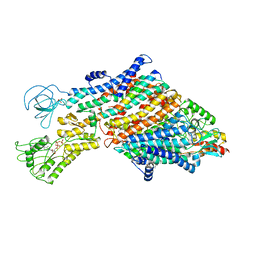 | | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Rheinberger, J, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRL
 
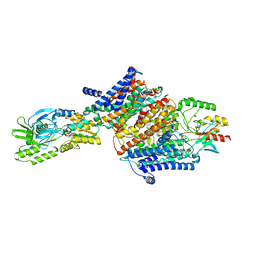 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E2-P conformation, under turnover conditions | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRE
 
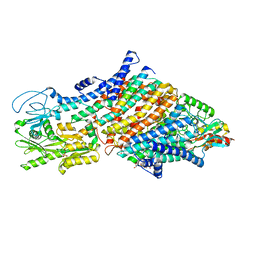 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, under turnover conditions | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRK
 
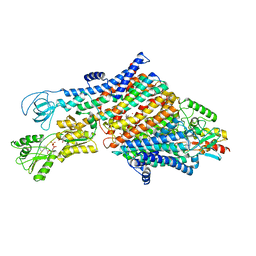 | | Cryo-EM map of the WT KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRH
 
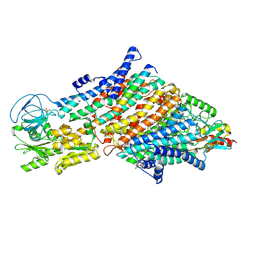 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
6FNW
 
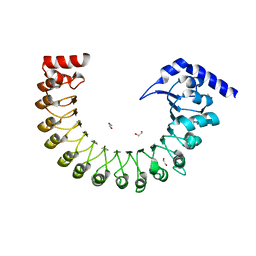 | | Structure of a volume-regulated anion channel of the LRRC8 family | | Descriptor: | 1,2-ETHANEDIOL, Volume-regulated anion channel subunit LRRC8A | | Authors: | Deneka, D, Sawicka, M, Lam, A.K.M, Paulino, C, Dutzler, R. | | Deposit date: | 2018-02-05 | | Release date: | 2018-05-16 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a volume-regulated anion channel of the LRRC8 family.
Nature, 558, 2018
|
|
6G9O
 
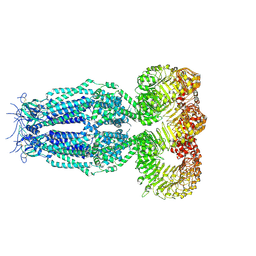 | | Structure of full-length homomeric mLRRC8A volume-regulated anion channel at 4.25 A resolution | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Sawicka, M, Deneka, D, Lam, A.K.M, Paulino, C, Dutzler, R. | | Deposit date: | 2018-04-11 | | Release date: | 2018-05-16 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Structure of a volume-regulated anion channel of the LRRC8 family.
Nature, 558, 2018
|
|
6G8Z
 
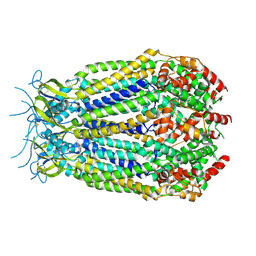 | | Structure of the pore domain of homomeric mLRRC8A volume-regulated anion channel at 3.66 A resolution | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Sawicka, M, Deneka, D, Lam, A.K.M, Paulino, C, Dutzler, R. | | Deposit date: | 2018-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure of a volume-regulated anion channel of the LRRC8 family.
Nature, 558, 2018
|
|
6G9L
 
 | | Structure of homomeric mLRRC8A volume-regulated anion channel at 5.01 A resolution | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Sawicka, M, Deneka, D, Lam, A.K.M, Paulino, C, Dutzler, R. | | Deposit date: | 2018-04-11 | | Release date: | 2018-05-16 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | Structure of a volume-regulated anion channel of the LRRC8 family.
Nature, 558, 2018
|
|
