6O47
 
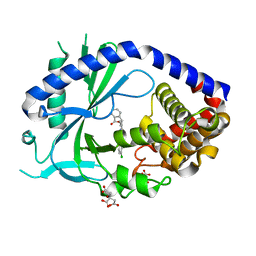 | | human cGAS core domain (K427E/K428E) bound with RU-521 | | Descriptor: | (3~{S})-3-[1-[4,5-bis(chloranyl)-1~{H}-benzimidazol-2-yl]-3-methyl-5-oxidanyl-pyrazol-4-yl]-3~{H}-2-benzofuran-1-one, 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, CITRIC ACID, ... | | Authors: | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O6Z
 
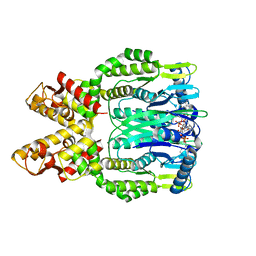 | | Crystal structure of Csm6 H381A in complex with cA4 by cocrystallization of cA4 and Csm6 | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6O79
 
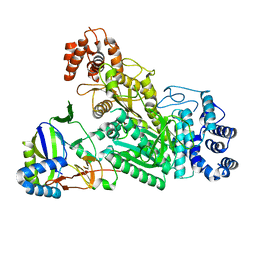 | | Crystal structure of Csm1-Csm4 cassette in complex with cA3 | | Descriptor: | CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, cyclic RNA cA3 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
6O7E
 
 | |
6O7I
 
 | |
6O6X
 
 | |
6O74
 
 | |
6O7D
 
 | | Crystal structure of Csm1-Csm4 cassette in complex with one ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
6O6S
 
 | | Crystal structure of Apo Csm6 | | Descriptor: | Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6O7H
 
 | |
6OV0
 
 | |
6PPR
 
 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP and DNA | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (70-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PPJ
 
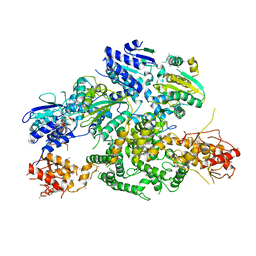 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), IRON/SULFUR CLUSTER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PPU
 
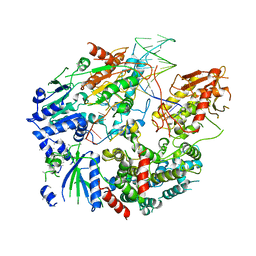 | | Cryo-EM structure of AdnAB-AMPPNP-DNA complex | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (29-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6UF1
 
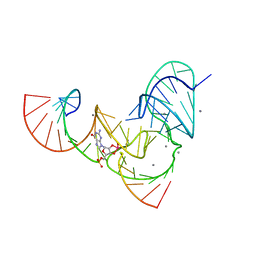 | | Pistol ribozyme transition-state analog vanadate | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(GVA)*UP*CP*CP*AP*A)-3'), RNA (50-MER) | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5BNX
 
 | |
6UFJ
 
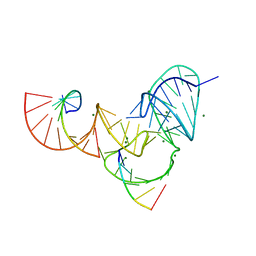 | | Pistol ribozyme product crystal structure | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*CP*AP*G)-3'), RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(23G))-3'), ... | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5BSA
 
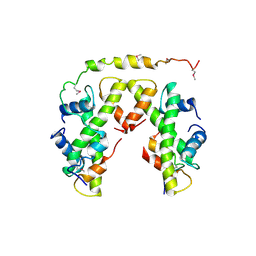 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.611 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
6UEY
 
 | | Pistol ribozyme transition-state analog vanadate | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(GVA)*UP*CP*CP*AP*A)-3'), RNA (50-MER) | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5BO0
 
 | |
6UFK
 
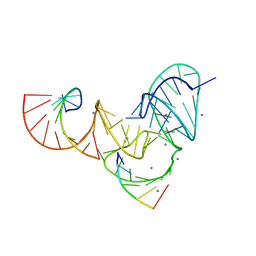 | | Pistol ribozyme product crystal soaked in Mn2+ | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*UP*CP*CP*AP*G)-3'), RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(23G))-3'), ... | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5BS7
 
 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog, ... | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
5BNV
 
 | |
6V9Q
 
 | | Cryo-EM structure of Cascade-TniQ binary complex | | Descriptor: | Cas8, RNA (61-MER), TniQ family protein, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-12-15 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-function insights into the initial step of DNA integration by a CRISPR-Cas-Transposon complex.
Cell Res., 30, 2020
|
|
6VBW
 
 | |
