7Y1W
 
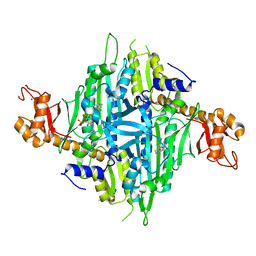 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | (2R,3S)-2-[3-[4,5-bis(chloranyl)benzimidazol-1-yl]propyl]piperidin-3-ol, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7Y28
 
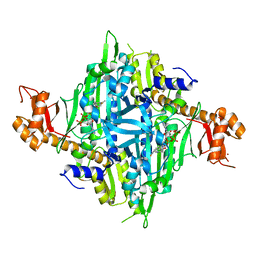 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | 1-[6-(3-fluorophenyl)benzimidazol-1-yl]-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7Y3S
 
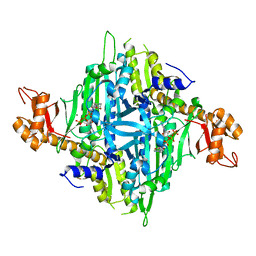 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | 1-(6-bromanyl-7-methyl-imidazo[4,5-b]pyridin-3-yl)-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-12 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
8JQV
 
 | |
1EHG
 
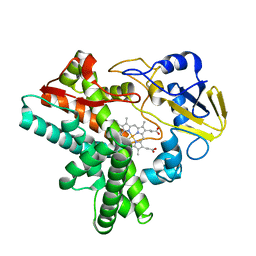 | |
1EHF
 
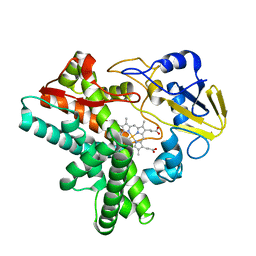 | |
1EHE
 
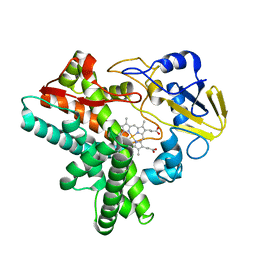 | |
7DP1
 
 | |
7DP0
 
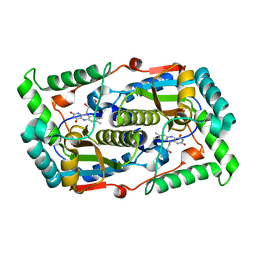 | |
7DP2
 
 | |
1CQS
 
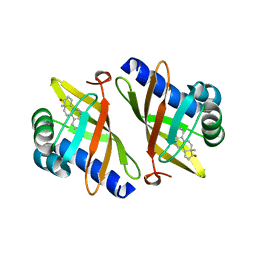 | | CRYSTAL STRUCTURE OF D103E MUTANT WITH EQUILENINEOF KSI IN PSEUDOMONAS PUTIDA | | Descriptor: | EQUILENIN, PROTEIN : KETOSTEROID ISOMERASE | | Authors: | Choi, G, Ha, N.C, Kim, S.W, Kim, D.H, Park, S, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-08-11 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Asp-99 donates a hydrogen bond not to Tyr-14 but to the steroid directly
in the catalytic mechanism of Delta 5-3-ketosteroid isomerase from
Pseudomonas putida biotype B
Biochemistry, 39, 2000
|
|
2PPO
 
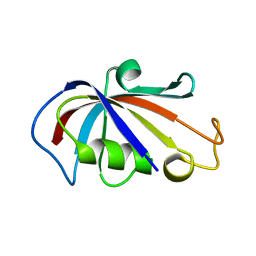 | |
2PPN
 
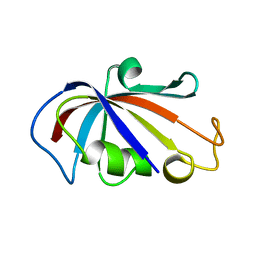 | |
2PPP
 
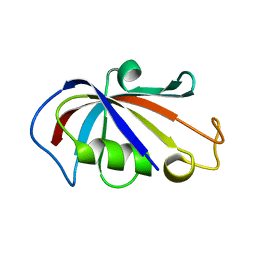 | |
4FZ5
 
 | | Crystal Structure of Human TIRAP TIR-domain | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Woo, J.R, Kim, S, Shoelson, S.E, Park, S. | | Deposit date: | 2012-07-06 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray Crystallographic Structure of TIR-Domain from the Human TIR-Domain Containing Adaptor Protein/MyD88 Adaptor-Like Protein (TIRAP/MAL)
Bull.Korean Chem.Soc., 33, 2013
|
|
2KYK
 
 | | The sandwich region between two LMP2A PY motif regulates the interaction between AIP4WW2domain and PY motif | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog | | Authors: | Seo, M, Park, S, Seok, S, Kim, J, Cha, M, Lee, B. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-01 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The sandwich region between two LMP2A PY motif regulates the interaction between AIP4WW2domain and PY motif
To be Published
|
|
1QKK
 
 | | Crystal structure of the receiver domain and linker region of DctD from Sinorhizobium meliloti | | Descriptor: | C4-DICARBOXYLATE TRANSPORT TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Meyer, M.G, Park, S, Zeringue, L, Staley, M, Mckinstry, M, Kaufman, R.I, Zhang, H, Yan, D, Yennawar, N, Farber, G.K, Nixon, B.T. | | Deposit date: | 1999-07-23 | | Release date: | 2000-07-30 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A dimeric two-component receiver domain inhibits the sigma54-dependent ATPase in DctD.
Faseb J., 15, 2001
|
|
4JNJ
 
 | | Structure based engineering of streptavidin monomer with a reduced biotin dissociation rate | | Descriptor: | BIOTIN, Streptavidin/Rhizavidin Hybrid, ZINC ION | | Authors: | DeMonte, D, Drake, E.J, Hong Lim, K, Gulick, A.M, Park, S. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structure-based engineering of streptavidin monomer with a reduced biotin dissociation rate.
Proteins, 81, 2013
|
|
4JOU
 
 | |
2AIV
 
 | | Multiple conformations in the ligand-binding site of the yeast nuclear pore targeting domain of NUP116P | | Descriptor: | fragment of Nucleoporin NUP116/NSP116 | | Authors: | Robinson, M.A, Park, S, Sun, Z.-Y.J, Silver, P, Wagner, G, Hogle, J. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Multiple Conformations in the Ligand-binding Site of the Yeast Nuclear Pore-targeting Domain of Nup116p
J.Biol.Chem., 280, 2005
|
|
2NC6
 
 | | Solution Structure of N-L-idosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-L-idopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
1J7N
 
 | | Anthrax Toxin Lethal factor | | Descriptor: | Lethal Factor precursor, SULFATE ION, ZINC ION | | Authors: | Pannifer, A.D, Wong, T.Y, Schwarzenbacher, R, Renatus, M, Petosa, C, Collier, R.J, Bienkowska, J, Lacy, D.B, Park, S, Leppla, S.H, Hanna, P, Liddington, R.C. | | Deposit date: | 2001-05-17 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anthrax lethal factor.
Nature, 414, 2001
|
|
2NC5
 
 | | Solution Structure of N-Xylosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-D-xylopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC3
 
 | | Solution Structure of N-Allosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-D-allopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
