5AWX
 
 | | Crystal structure of Human PTPRZ D1 domain | | Descriptor: | BROMIDE ION, Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Sugawara, H. | | Deposit date: | 2015-07-10 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Small-molecule inhibition of PTPRZ reduces tumor growth in a rat model of glioblastoma
Sci Rep, 6, 2016
|
|
4DBG
 
 | | Crystal structure of HOIL-1L-UBL complexed with a HOIP-UBA derivative | | Descriptor: | RING finger protein 31, RanBP-type and C3HC4-type zinc finger-containing protein 1 | | Authors: | Yagi, H, Hiromoto, T, Mizushima, T, Kurimoto, E, Kato, K. | | Deposit date: | 2012-01-15 | | Release date: | 2012-04-04 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A non-canonical UBA-UBL interaction forms the linear-ubiquitin-chain assembly complex
Embo Rep., 13, 2012
|
|
2RQP
 
 | |
6LE4
 
 | |
6LDO
 
 | |
7JRI
 
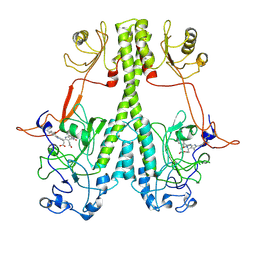 | |
7JR5
 
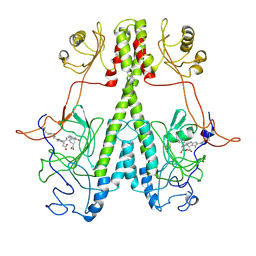 | | Real Time Reaction Intermediates in Stigmatella Bacteriophytochrome P2 | | Descriptor: | 3-[2-[[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-5-[[(3~{S})-4-ethyl-3-methyl-2-oxidanylidene-1,3-dihydropyrrol-5-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, BENZAMIDINE, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structures of transient intermediates in the phytochrome photocycle.
Structure, 29, 2021
|
|
5X7Y
 
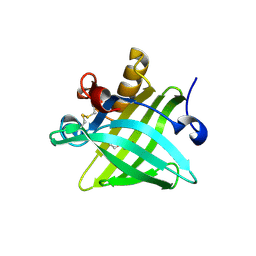 | | Crystal Structure of the Dog Lipocalin Allergen Can f 6 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipocalin-Can f 6 allergen | | Authors: | Yamamoto, K, Otani, T, Sugiura, K, Nakatsuji, M, Nishimura, S, Inui, T. | | Deposit date: | 2017-02-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the dog allergen Can f 6 and structure-based implications of its cross-reactivity with the cat allergen Fel d 4.
Sci Rep, 9, 2019
|
|
5Z30
 
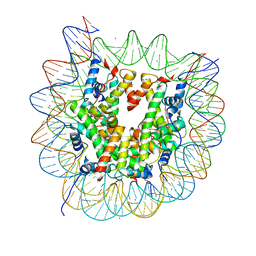 | | The crystal structure of the nucleosome containing a cancer-associated histone H2A.Z R80C mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.Z, ... | | Authors: | Horikoshi, N, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2018-01-05 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
5B1H
 
 | |
5B1I
 
 | |
3WSV
 
 | |
3WSW
 
 | |
5DSV
 
 | | Crystal structure of human proteasome alpha7 tetradecamer | | Descriptor: | Proteasome subunit alpha type-3 | | Authors: | Satoh, T, Thammaporn, R, Seetaha, S, Kato, K. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Disassembly of the self-assembled, double-ring structure of proteasome alpha 7 homo-tetradecamer by alpha 6
Sci Rep, 5, 2015
|
|
3AZO
 
 | | Crystal structure of puromycin hydrolase | | Descriptor: | Aminopeptidase, SULFATE ION | | Authors: | Matoba, Y, Sugiyama, M. | | Deposit date: | 2011-05-27 | | Release date: | 2011-07-27 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence that puromycin hydrolase is a new type of aminopeptidase with a prolyl oligopeptidase family fold
Proteins, 79, 2011
|
|
3AZP
 
 | |
3AZQ
 
 | |
3WA6
 
 | |
3WA7
 
 | | Crystal structure of selenomethionine-labeled tannase from Lactobacillus plantarum in the orthorhombic crystal | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Matoba, Y, Tanaka, N, Sugiyama, M. | | Deposit date: | 2013-04-27 | | Release date: | 2013-07-24 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic and mutational analyses of tannase from Lactobacillus plantarum.
Proteins, 81, 2013
|
|
2ZWF
 
 | |
2ZWE
 
 | |
2ZMZ
 
 | |
2ZMY
 
 | |
2ZWD
 
 | |
2ZWG
 
 | |
