5BKB
 
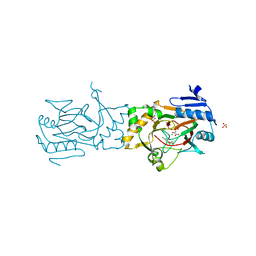 | | Crystal structure of AAD-1 in complex with (R)-dichlorprop, Mn(II), and 2-oxoglutarate | | Descriptor: | (2R)-2-(2,4-dichlorophenoxy)propanoic acid, (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, 2-OXOGLUTARIC ACID, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2019-06-02 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5BST
 
 | |
5BSU
 
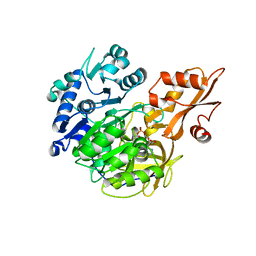 | | Crystal structure of 4-coumarate:CoA ligase complexed with caffeoyl adenylate | | Descriptor: | 4-coumarate--CoA ligase 2, 5'-O-[(R)-{[(2E)-3-(3,4-dioxocyclohexa-1,5-dien-1-yl)prop-2-enoyl]oxy}(hydroxy)phosphoryl]adenosine, GLYCEROL, ... | | Authors: | Li, Z, Nair, S.K. | | Deposit date: | 2015-06-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Specificity and Flexibility in a Plant 4-Coumarate:CoA Ligase.
Structure, 23, 2015
|
|
4E5P
 
 | |
4EBF
 
 | |
4E5K
 
 | |
4E5M
 
 | |
4E5N
 
 | | Thermostable phosphite dehydrogenase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Thermostable phosphite dehydrogenase | | Authors: | Zou, Y, Zhang, H, Nair, S.K. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of phosphite dehydrogenase provide insights into nicotinamide cofactor regeneration.
Biochemistry, 51, 2012
|
|
5DLY
 
 | |
5DM1
 
 | |
5DM0
 
 | | Crystal structure of the plantazolicin methyltransferase BamL in complex with triazolic desmethylPZN analog and SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ethyl 2-(2-{2-[(1S)-1-amino-4-carbamimidamidobutyl]-1,3-thiazol-4-yl}-5-methyl-1,3-oxazol-4-yl)-1,3-thiazole-4-carboxylate, ... | | Authors: | Hao, Y, Nair, S.K. | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into methyltransferase specificity and bioactivity of derivatives of the antibiotic plantazolicin.
Acs Chem.Biol., 10, 2015
|
|
5DM4
 
 | | Crystal structure of the plantazolicin methyltransferase BpumL in complex with pentazolic desmethylPZN analog and SAH | | Descriptor: | 1-[(4S)-4-(4-{4-[4-(5,5'-dimethyl-2,4'-bi-1,3-oxazol-2'-yl)-1,3-thiazol-2-yl]-5-methyl-1,3-oxazol-2-yl}-1,3-thiazol-2-yl)-4-(methylamino)butyl]guanidine, GLYCEROL, Methyltransferase domain family, ... | | Authors: | Hao, Y, Nair, S.K. | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into methyltransferase specificity and bioactivity of derivatives of the antibiotic plantazolicin.
Acs Chem.Biol., 10, 2015
|
|
5DM2
 
 | |
5DZT
 
 | |
5EHK
 
 | |
5F2K
 
 | |
5F2O
 
 | |
5F2N
 
 | |
6C2S
 
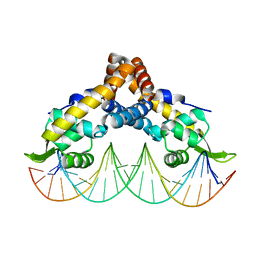 | | Transcriptional repressor, CouR, bound to a 23-mer DNA duplex | | Descriptor: | 23-mer, Transcriptional regulator, MarR family | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-08 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
6C8R
 
 | |
6CIB
 
 | |
6CI7
 
 | |
6CGQ
 
 | | Threonine synthase from Bacillus subtilis ATCC 6633 with PLP and PLP-Ala | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Petronikolou, N, Nair, S.K. | | Deposit date: | 2018-02-20 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Molecular Basis of Bacillus subtilis ATCC 6633 Self-Resistance to the Phosphono-oligopeptide Antibiotic Rhizocticin.
ACS Chem. Biol., 14, 2019
|
|
6D6D
 
 | |
6D6O
 
 | |
