1EGF
 
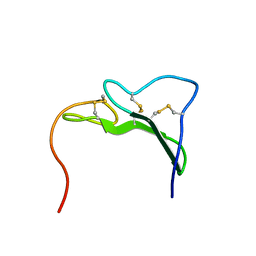 | |
3EGF
 
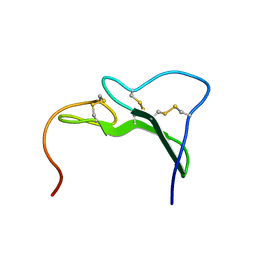 | |
1NS1
 
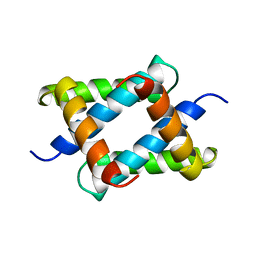 | |
2SPZ
 
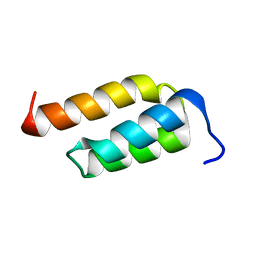 | | STAPHYLOCOCCAL PROTEIN A, Z-DOMAIN, NMR, 10 STRUCTURES | | 分子名称: | IMMUNOGLOBULIN G BINDING PROTEIN A | | 著者 | Montelione, G.T, Tashiro, M, Tejero, R, Lyons, B.A. | | 登録日 | 1998-07-29 | | 公開日 | 1998-08-05 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-resolution solution NMR structure of the Z domain of staphylococcal protein A.
J.Mol.Biol., 272, 1997
|
|
1AIL
 
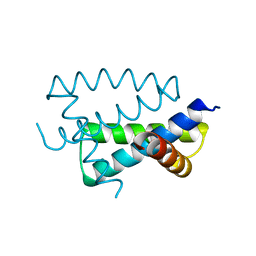 | | N-TERMINAL FRAGMENT OF NS1 PROTEIN FROM INFLUENZA A VIRUS | | 分子名称: | NONSTRUCTURAL PROTEIN NS1 | | 著者 | Liu, J, Lynch, P.A, Chien, C, Montelione, G.T, Krug, R.M, Berman, H.M. | | 登録日 | 1997-04-21 | | 公開日 | 1997-10-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein.
Nat.Struct.Biol., 4, 1997
|
|
4ZEQ
 
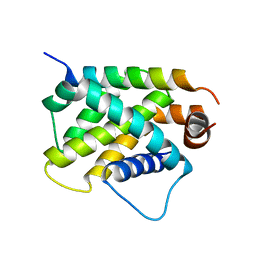 | | Crystal Structure of human BFL-1 in complex with tBid BH3 peptide, Northeast Structural Genomics Consortium Target HX9247 | | 分子名称: | BH3-interacting domain death agonist, Bcl-2-related protein A1 | | 著者 | Guan, R, Xiao, R, Mao, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-04-20 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of human BFL-1 in complex with tBid BH3 peptide
To Be Published
|
|
8CWA
 
 | |
8CTO
 
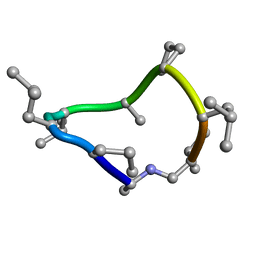 | |
1KKG
 
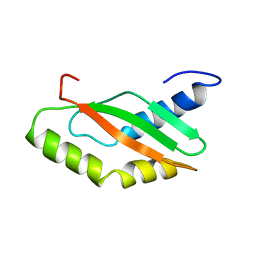 | | NMR Structure of Ribosome-Binding Factor A (RbfA) | | 分子名称: | ribosome-binding factor A | | 著者 | Huang, Y.J, Swapna, G.V.T, Rajan, P.K, Ke, H, Xia, B, Shukla, K, Inouye, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2001-12-07 | | 公開日 | 2003-03-18 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of Ribosome-binding Factor A (RbfA), A Cold-shock
Adaptation Protein from Escherichia coli
J.Mol.Biol., 327, 2003
|
|
8CUN
 
 | |
7K3S
 
 | |
5DIL
 
 | | Crystal structure of the effector domain of the NS1 protein from influenza virus B | | 分子名称: | IODIDE ION, Non-structural protein 1 | | 著者 | Guan, R, Hamilton, K, Ma, L, Montelione, G.T. | | 登録日 | 2015-09-01 | | 公開日 | 2016-08-10 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | A Second RNA-Binding Site in the NS1 Protein of Influenza B Virus.
Structure, 24, 2016
|
|
5GAJ
 
 | | Solution NMR structure of De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258 | | 分子名称: | DE NOVO DESIGNED PROTEIN OR258 | | 著者 | Liu, G, Castelllanos, J, Koga, R, Koga, N, Xiao, R, Pederson, K, Janjua, H, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-12-01 | | 公開日 | 2016-01-06 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258
To Be Published
|
|
6XEH
 
 | |
7JQ8
 
 | | Solution NMR structure of human Brd3 ET domain | | 分子名称: | Bromodomain-containing protein 3, Integrase peptide | | 著者 | Aiyer, S, Swapna, G.V.T, Roth, J.M, Montelione, G.T. | | 登録日 | 2020-08-10 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
6MSP
 
 | | De novo Designed Protein Foldit3 | | 分子名称: | De novo Designed Protein Foldit3 | | 著者 | Liu, G, Ishida, Y, Swapna, G.V.T, Kleinfelter, S, Koepnick, B, Baker, D, Montelione, G.T. | | 登録日 | 2018-10-17 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
8CDC
 
 | | Native 3CLpro from SARS-CoV-2 at 1.54 A | | 分子名称: | 3C-like proteinase | | 著者 | Mazzei, L, Jovanovic, A, Acton, T.B, Ciurli, S, Montelione, G.T. | | 登録日 | 2023-01-30 | | 公開日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Expression, purification, and characterization of SARS-CoV2 3CLpro in a mature form
To Be Published
|
|
5KPH
 
 | |
5KPE
 
 | | Solution NMR Structure of Denovo Beta Sheet Design Protein, Northeast Structural Genomics Consortium (NESG) Target OR664 | | 分子名称: | De novo Beta Sheet Design Protein OR664 | | 著者 | Tang, Y, Liu, G, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2016-07-03 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
7DLM
 
 | | Short chain dehydrogenase (SCR) crystal structure with NADPH | | 分子名称: | Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-11-28 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLL
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADPH | | 分子名称: | (S)-specific carbonyl reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-11-28 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DN1
 
 | | Hetero-oligomers of SCR-SCR2 crystal structure with NADPH | | 分子名称: | (S)-specific carbonyl reductase, Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-12-08 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DMG
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADP | | 分子名称: | (S)-specific carbonyl reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-12-03 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLD
 
 | | Crystal structures of (S)-carbonyl reductases from Candida parapsilosis in different oligomerization states | | 分子名称: | Carbonyl Reductase, MAGNESIUM ION | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-11-27 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7KBQ
 
 | |
