5GN8
 
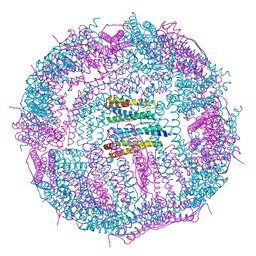 | | Structure of a 48-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | CALCIUM ION, Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Zhao, G, Mikami, B. | | Deposit date: | 2016-07-19 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | "Silent" Amino Acid Residues at Key Subunit Interfaces Regulate the Geometry of Protein Nanocages
ACS Nano, 10, 2016
|
|
5GX7
 
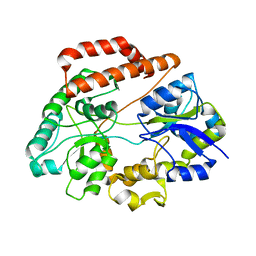 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with a sulfate group at C-6 position of GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, CALCIUM ION, Extracellular solute-binding protein family 1 | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-09-15 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
5GUB
 
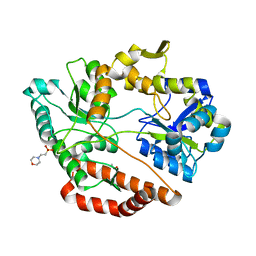 | | Crystal structure of solute-binding protein complexed with sulfate group-free unsaturated chondroitin disaccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-08-26 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
5GX8
 
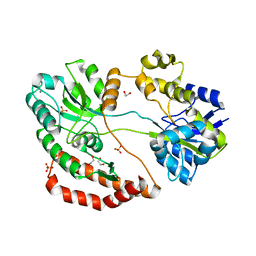 | | Crystal structure of solute-binding protein related to glycosaminoglycans | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-09-15 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
5GX6
 
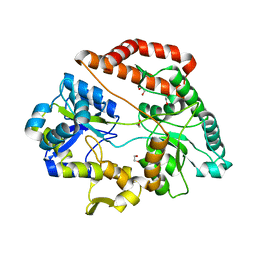 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with a sulfate group at C-4 position of GalNAc | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-09-15 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
3GNE
 
 | | Crystal structure of alginate lyase vAL-1 from Chlorella virus | | Descriptor: | CITRATE ANION, GLYCEROL, VAL-1 | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-03-17 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
5H71
 
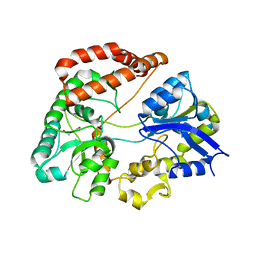 | | Structure of alginate-binding protein AlgQ2 in complex with an alginate trisaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, AlgQ2, CALCIUM ION, ... | | Authors: | Uenishi, K, Kaneko, A, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-11-15 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate
J. Biol. Chem., 292, 2017
|
|
5HXI
 
 | | 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5HN bound | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Mikami, B. | | Deposit date: | 2016-01-30 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of the Tyr270 residue in 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti
J. Biosci. Bioeng., 123, 2017
|
|
5H6U
 
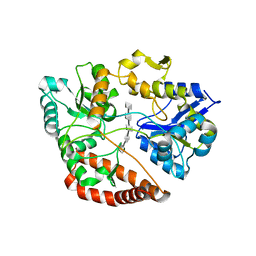 | | Structure of alginate-binding protein AlgQ2 in complex with an alginate pentasaccharide | | Descriptor: | AlgQ2, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Uenishi, K, Kaneko, A, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-11-15 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate
J. Biol. Chem., 292, 2017
|
|
7CPK
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
7CPL
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
7C9J
 
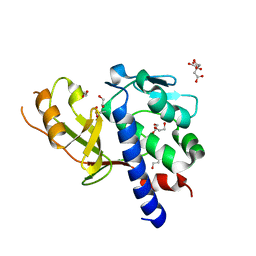 | | Transglutaminase from Geobacillus stearothermophilus (without C-terminal extension) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Protein-glutamine gamma-glutamyltransferase | | Authors: | Takita, T, Mikami, B, Lei, Y, Jing, Y, Yamada, A, Yasukawa, K. | | Deposit date: | 2020-06-06 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transglutaminase from Geobacillus stearothermophilus (without C-terminal extension)
To be published
|
|
3IM0
 
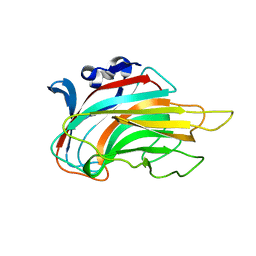 | | Crystal structure of Chlorella virus vAL-1 soaked in 200mM D-glucuronic acid, 10% PEG-3350, and 200mM glycine-NaOH (pH 10.0) | | Descriptor: | VAL-1, beta-D-glucopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-08-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
7EKD
 
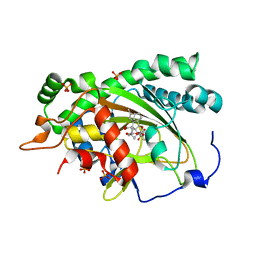 | | Crystal structure of gibberellin 3-oxidase 2 (GA3ox2) in rice | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-OXOGLUTARIC ACID, Gibberellin 3-beta-dioxygenase 2, ... | | Authors: | Takehara, S, Kawai, K, Mikami, B, Ueguchi-Tanaka, M. | | Deposit date: | 2021-04-05 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Evolutionary alterations in gene expression and enzymatic activities of gibberellin 3-oxidase 1 in Oryza.
Commun Biol, 5, 2022
|
|
7E4S
 
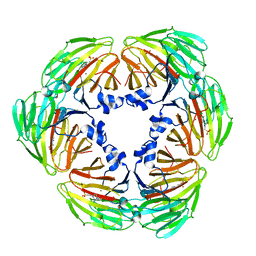 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-dehydro-4-deoxy-D-glucuronate isomerase, ZINC ION | | Authors: | Yamamoto, Y, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-02-15 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structures of Lacticaseibacillus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI in complex with substrate analogs
J.Appl.Glyosci., 2023
|
|
7E8R
 
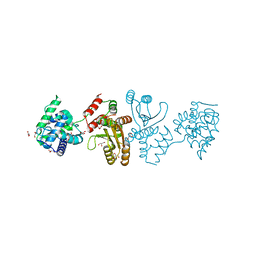 | | EcoT38I restriction endonuclease | | Descriptor: | EcoT38I restriction endonuclease, GLYCEROL | | Authors: | Kita, K, Mikami, B. | | Deposit date: | 2021-03-02 | | Release date: | 2022-03-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of EcoT38I restriction endonuclease
To Be Published
|
|
3K3T
 
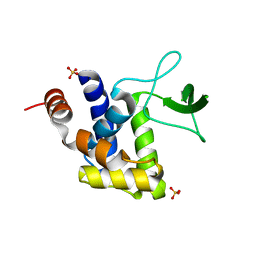 | | E185A mutant of peptidoglycan hydrolase from Sphingomonas sp. A1 | | Descriptor: | Peptidoglycan hydrolase FlgJ, SULFATE ION | | Authors: | Maruyama, Y, Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-10-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mutational studies of the peptidoglycan hydrolase FlgJ of Sphingomonas sp. strain A1
J.Basic Microbiol., 50, 2010
|
|
7EDB
 
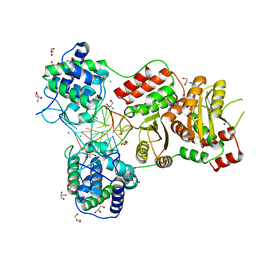 | | EcoT38I restriction endonuclease complexed with DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kita, K, Mikami, B. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural analysis of EcoT38I restriction endonuclease
To Be Published
|
|
3KSC
 
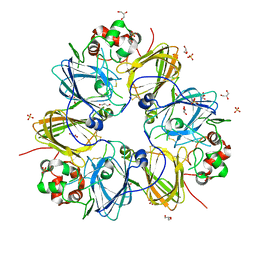 | | Crystal structure of pea prolegumin, an 11S seed globulin from Pisum sativum L. | | Descriptor: | GLYCEROL, LegA class, SULFATE ION | | Authors: | Tandang-Silvas, M.R.G, Fukuda, T, Fukuda, C, Prak, K, Cabanos, C, Kimura, A, Itoh, T, Mikami, B, Maruyama, N, Utsumi, S. | | Deposit date: | 2009-11-21 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KGL
 
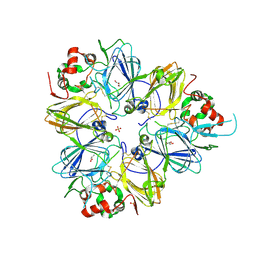 | | Crystal structure of procruciferin, 11S globulin from Brassica napus | | Descriptor: | Cruciferin, GLYCEROL, SULFATE ION | | Authors: | Tandang-Silvas, M.R, Mikami, B, Maruyama, N, Utsumi, S. | | Deposit date: | 2009-10-29 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 1804, 2010
|
|
1HV6
 
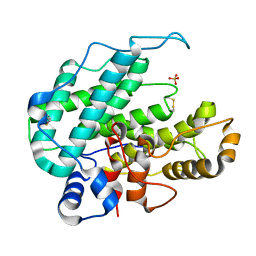 | | CRYSTAL STRUCTURE OF ALGINATE LYASE A1-III COMPLEXED WITH TRISACCHARIDE PRODUCT. | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-D-glucopyranuronic acid, ALGINATE LYASE, SULFATE ION | | Authors: | Yoon, H.-J, Hashimoto, W, Miyake, O, Murata, K, Mikami, B. | | Deposit date: | 2001-01-08 | | Release date: | 2001-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 A resolution.
J.Mol.Biol., 307, 2001
|
|
1J1N
 
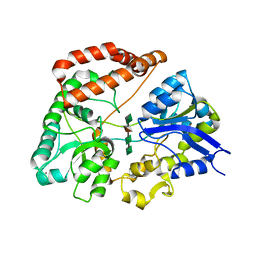 | | Structure Analysis of AlgQ2, A Macromolecule(Alginate)-Binding Periplasmic Protein Of Sphingomonas Sp. A1., Complexed with an Alginate Tetrasaccharide | | Descriptor: | AlgQ2, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mikami, B, Mishima, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2002-12-11 | | Release date: | 2003-06-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of AlgQ2, a macromolecule (alginate)-binding protein of Sphingomonas sp. A1, complexed with an alginate tetrasaccharide at 1.6-A resolution
J.BIOL.CHEM., 278, 2003
|
|
5SW0
 
 | |
5SW1
 
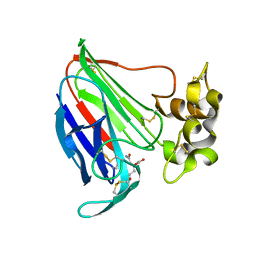 | | Thaumatin Structure at pH 6.0 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Thaumatin Structure at pH 6.0
To Be Published
|
|
5SW2
 
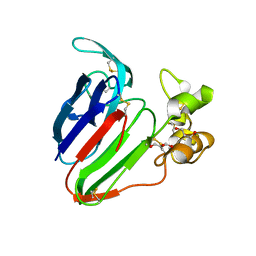 | | Thaumatin Structure at pH 6.0, orthorhombic type1 | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Thaumatin Structure at pH 6.0, orthorhombic type1
To Be Published
|
|
