3MEY
 
 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bll0957 protein, MAGNESIUM ION, ... | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MF1
 
 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with an analogue of glycyl adenylate | | Descriptor: | 5'-O-(glycylsulfamoyl)adenosine, Bll0957 protein, ZINC ION | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7PVO
 
 | | Adenylosuccinate Synthetase from H. pylori in complex with IMP | | Descriptor: | Adenylosuccinate synthetase, INOSINIC ACID, SULFATE ION | | Authors: | Narczyk, M, Bzowska, A, Maksymiuk, W. | | Deposit date: | 2021-10-05 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pursuit of new alternative ways to eradicate Helicobacter pylori continues: Detailed characterization of interactions in the adenylosuccinate synthetase active site.
Int.J.Biol.Macromol., 226, 2023
|
|
6ZXQ
 
 | | Adenylosuccinate Synthetase from H. pylori in complex with HDA, GDP, IMO, Mg | | Descriptor: | 6-O-PHOSPHORYL INOSINE MONOPHOSPHATE, Adenylosuccinate synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bubic, A, Stefanic, Z. | | Deposit date: | 2020-07-30 | | Release date: | 2021-08-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The pursuit of new alternative ways to eradicate Helicobacter pylori continues: Detailed characterization of interactions in the adenylosuccinate synthetase active site
Int.J.Biol.Macromol., 2023
|
|
3UT6
 
 | | Crystal structure of E. Coli PNP complexed with PO4 and formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2011-11-25 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | New phosphate binding sites in the crystal structure of Escherichia coli purine nucleoside phosphorylase complexed with phosphate and formycin A.
Febs Lett., 586, 2012
|
|
1VFN
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | HYPOXANTHINE, MAGNESIUM ION, PURINE-NUCLEOSIDE PHOSPHORYLASE, ... | | Authors: | Koellner, G, Bzowska, A. | | Deposit date: | 1996-10-08 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calf spleen purine nucleoside phosphorylase in a complex with hypoxanthine at 2.15 A resolution.
J.Mol.Biol., 265, 1997
|
|
7OOY
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzylthio-2-chloropurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzylthio-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7OOZ
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzyloxo-2-chloropurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzyloxo-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7OPA
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzylthiopurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzylthio-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7OP9
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 2,6-dichloropurine | | Descriptor: | 2,6-bis(chloranyl)-7H-purine, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
4TS9
 
 | |
4TTJ
 
 | | Crystal structure of double mutant E. Coli purine nucleoside phosphorylase with 6 FMC molecules | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type, ... | | Authors: | Stefanic, Z, Bzowska, A. | | Deposit date: | 2014-06-21 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Crystallographic snapshots of ligand binding to hexameric purine nucleoside phosphorylase and kinetic studies give insight into the mechanism of catalysis.
Sci Rep, 8, 2018
|
|
4TTA
 
 | | Crystal structure of double mutant E. Coli purine nucleoside phosphorylase with 2 FMC molecules | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type, ... | | Authors: | Stefanic, Z, Bzowska, A. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic snapshots of ligand binding to hexameric purine nucleoside phosphorylase and kinetic studies give insight into the mechanism of catalysis.
Sci Rep, 8, 2018
|
|
4TTI
 
 | | Crystal structure of double mutant E. Coli purine nucleoside phosphorylase with 4 FMC molecules | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type, ... | | Authors: | Stefanic, Z, Bzowska, A. | | Deposit date: | 2014-06-21 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallographic snapshots of ligand binding to hexameric purine nucleoside phosphorylase and kinetic studies give insight into the mechanism of catalysis.
Sci Rep, 8, 2018
|
|
4TS3
 
 | |
5LU0
 
 | | Crystal structure of H. pylori referent strain in complex with PO4 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Stefanic, Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
6F52
 
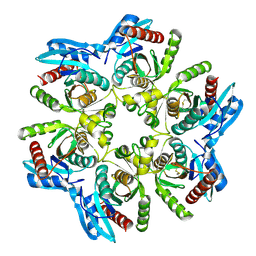 | |
6F4W
 
 | | Crystal structure of H. pylori purine nucleoside phosphorylase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, Purine nucleoside phosphorylase DeoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
6F5I
 
 | | Crystal structure of H. pylori purine nucleoside phosphorylase | | Descriptor: | METHANOL, Purine nucleoside phosphorylase DeoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
6F5A
 
 | | Crystal structure of H. pylori purine nucleoside phosphorylase | | Descriptor: | Purine nucleoside phosphorylase DeoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
6F4X
 
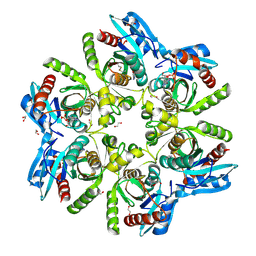 | |
1LVU
 
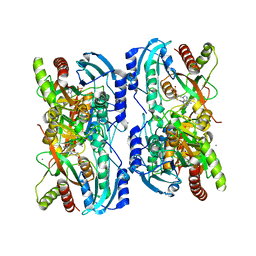 | | Crystal structure of calf spleen purine nucleoside phosphorylase in a new space group with full trimer in the asymmetric unit | | Descriptor: | 2,6-DIAMINO-(S)-9-[2-(PHOSPHONOMETHOXY)PROPYL]PURINE, CALCIUM ION, Purine nucleoside phosphorylase | | Authors: | Bzowska, A, Koellner, G, Wielgus-Kutrowska, B, Stroh, A, Raszewski, G, Holy, A, Steiner, T, Frank, J. | | Deposit date: | 2002-05-29 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of calf spleen purine nucleoside phosphorylase with two full trimers in the asymmetric unit: important implications for the mechanism of catalysis
J.Mol.Biol., 342, 2004
|
|
1LV8
 
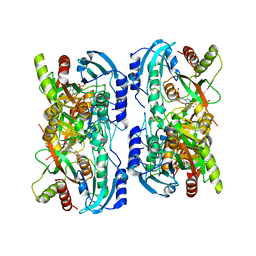 | | Crystal structure of calf spleen purine nucleoside phosphorylase in a new space group with full trimer in the asymmetric unit | | Descriptor: | 2,6-DIAMINO-(S)-9-[2-(PHOSPHONOMETHOXY)PROPYL]PURINE, CALCIUM ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Bzowska, A, Koellner, G, Wielgus-Kutrowska, B, Stroh, A, Raszewski, G, Holy, A, Steiner, T, Frank, J. | | Deposit date: | 2002-05-27 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of calf spleen purine nucleoside phosphorylase with two full trimers in the asymmetric unit: important implications for the mechanism of catalysis
J.Mol.Biol., 342, 2004
|
|
5MX6
 
 | |
5MX4
 
 | |
