2L0K
 
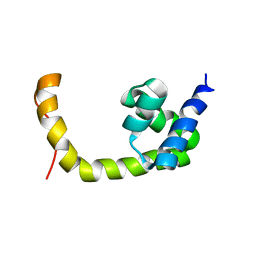 | | NMR solution structure of a transcription factor SpoIIID in complex with DNA | | 分子名称: | Stage III sporulation protein D | | 著者 | Chen, B, Himes, P, Lu, Z, Liu, A, Yan, H, Kroos, L. | | 登録日 | 2010-07-08 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Novel Mode of DNA Binding by Bacterial Transcription Factor SpoIIID
To be Published
|
|
3FZ2
 
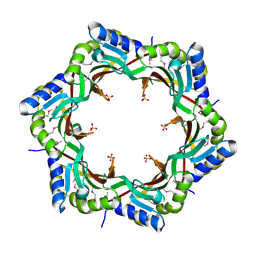 | | Crystal structure of the tail terminator protein from phage lambda (gpU-D74A) | | 分子名称: | Minor tail protein U, SULFATE ION | | 著者 | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | 登録日 | 2009-01-23 | | 公開日 | 2009-05-26 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
3FZB
 
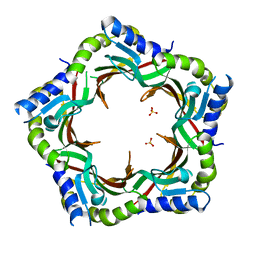 | | Crystal structure of the tail terminator protein from phage lambda (gpU-WT) | | 分子名称: | Minor tail protein U, SULFATE ION | | 著者 | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | 登録日 | 2009-01-24 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
6BVP
 
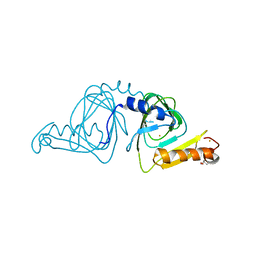 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase N27A from Cupriavidus metallidurans | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2017-12-13 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.903 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6BVQ
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase N27A from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2017-12-13 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.084 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6BVR
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142A from Cupriavidus metallidurans | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2017-12-13 | | 公開日 | 2018-06-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6BVS
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142A from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | 著者 | Yang, Y, Liu, F, Liu, A. | | 登録日 | 2017-12-13 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.318 Å) | | 主引用文献 | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6MGT
 
 | | Crystal structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase Mutant H110A | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | 著者 | Yang, Y, Daivs, I, Matsui, T, Rubalcava, I, Liu, A. | | 登録日 | 2018-09-14 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Quaternary structure of alpha-amino-beta-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) controls its activity.
J.Biol.Chem., 294, 2019
|
|
6MGS
 
 | | Crystal structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase with Space Group of C2221 | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | 著者 | Yang, Y, Davis, I, Matsui, T, Rubalcava, I, Liu, A. | | 登録日 | 2018-09-14 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.131 Å) | | 主引用文献 | Quaternary structure of alpha-amino-beta-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) controls its activity.
J.Biol.Chem., 294, 2019
|
|
4R52
 
 | | 1.5 angstrom crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | 著者 | Geng, J, Gumpper, R.H, Huo, L, Liu, A. | | 登録日 | 2014-08-20 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | 1.5 angstrom crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans
To be Published
|
|
4ERG
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, FE (III) ION | | 著者 | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | 登録日 | 2012-04-20 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.789 Å) | | 主引用文献 | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4ERI
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | 著者 | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | 登録日 | 2012-04-20 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.0006 Å) | | 主引用文献 | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4ERA
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | 著者 | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | 登録日 | 2012-04-19 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.398 Å) | | 主引用文献 | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4EPK
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | 著者 | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | 登録日 | 2012-04-17 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6009 Å) | | 主引用文献 | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4IFR
 
 | | 2.40 Angstroms X-ray crystal structure of R239A 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase from Pseudomonas fluorescens | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, ZINC ION | | 著者 | Huo, L, Davis, I, Chen, L, Liu, A. | | 登録日 | 2012-12-14 | | 公開日 | 2013-09-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.391 Å) | | 主引用文献 | The power of two: arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase.
J.Biol.Chem., 288, 2013
|
|
4IFK
 
 | | Arginines 51 and 239* from a Neighboring Subunit are Essential for Catalysis in a Zinc-dependent Decarboxylase | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | 著者 | Huo, L, Davis, I, Chen, L, Liu, A. | | 登録日 | 2012-12-14 | | 公開日 | 2013-09-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.012 Å) | | 主引用文献 | The power of two: arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase.
J.Biol.Chem., 288, 2013
|
|
4IFO
 
 | | 2.50 Angstroms X-ray crystal structure of R51A 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase from Pseudomonas fluorescens | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, ZINC ION | | 著者 | Huo, L, Davis, I, Chen, L, Liu, A. | | 登録日 | 2012-12-14 | | 公開日 | 2013-09-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The power of two: arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase.
J.Biol.Chem., 288, 2013
|
|
4IG2
 
 | | 1.80 Angstroms X-ray crystal structure of R51A and R239A heterodimer 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase from Pseudomonas fluorescens | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, ZINC ION | | 著者 | Huo, L, Davis, I, Chen, L, Liu, A. | | 登録日 | 2012-12-15 | | 公開日 | 2013-09-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The power of two: arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase.
J.Biol.Chem., 288, 2013
|
|
6AVJ
 
 | | Crystal structure of human Mitochondrial inner NEET protein (MiNT)/CISD3 | | 分子名称: | CDGSH iron-sulfur domain-containing protein 3, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | 著者 | Lipper, C.H, Karmi, O, Sohn, Y.S, Darash-Yahana, M, Lammert, H, Song, L, Liu, A, Mittler, R, Nechushtai, R, Onuchic, J.N, Jennings, P.A. | | 登録日 | 2017-09-02 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the human monomeric NEET protein MiNT and its role in regulating iron and reactive oxygen species in cancer cells.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6XP8
 
 | |
5OVQ
 
 | |
1Z1Z
 
 | |
6L42
 
 | |
1TMB
 
 | | MOLECULAR BASIS FOR THE INHIBITION OF HUMAN ALPHA-THROMBIN BY THE MACROCYCLIC PEPTIDE CYCLOTHEONAMIDE A | | 分子名称: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUGEN, ... | | 著者 | Qiu, X, Padmanabhan, K.P, Maryanoff, B.E, Tulinsky, A. | | 登録日 | 1993-05-27 | | 公開日 | 1994-01-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular basis for the inhibition of human alpha-thrombin by the macrocyclic peptide cyclotheonamide A.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
6PEU
 
 | |
