7YPG
 
 | |
3HU2
 
 | | Structure of p97 N-D1 R86A mutant in complex with ATPgS | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | 著者 | Tang, W.-K. | | 登録日 | 2009-06-12 | | 公開日 | 2010-06-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
3HU1
 
 | | Structure of p97 N-D1 R95G mutant in complex with ATPgS | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | 著者 | Tang, W.-K. | | 登録日 | 2009-06-12 | | 公開日 | 2010-06-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
5F6L
 
 | | The crystal structure of MLL1 (N3861I/Q3867L) in complex with RbBP5 and Ash2L | | 分子名称: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Li, Y, Lei, M, Chen, Y. | | 登録日 | 2015-12-06 | | 公開日 | 2016-02-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
3HU3
 
 | | Structure of p97 N-D1 R155H mutant in complex with ATPgS | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | 著者 | Tang, W.-K. | | 登録日 | 2009-06-12 | | 公開日 | 2010-06-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
5F59
 
 | | The crystal structure of MLL3 SET domain | | 分子名称: | Histone-lysine N-methyltransferase 2C, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | 著者 | Li, Y, Lei, M, Chen, Y. | | 登録日 | 2015-12-04 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F5E
 
 | |
5F6K
 
 | | Crystal structure of the MLL3-Ash2L-RbBP5 complex | | 分子名称: | Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Li, Y, Lei, M, Chen, Y. | | 登録日 | 2015-12-06 | | 公開日 | 2016-02-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.411 Å) | | 主引用文献 | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
7R1U
 
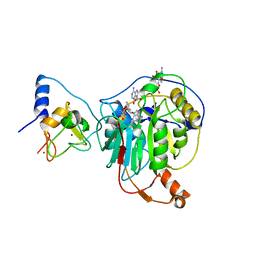 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | 分子名称: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | 登録日 | 2022-02-03 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7R1T
 
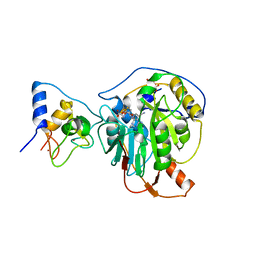 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | 分子名称: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | 登録日 | 2022-02-03 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
6KXW
 
 | | Crystal structure of human aquaporin AQP7 in bound to glycerol | | 分子名称: | Aquaporin-7, GLYCEROL | | 著者 | Zhang, L, Yao, D, Zhou, F, Zhang, Q, Zhou, L, Cao, Y. | | 登録日 | 2019-09-13 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | The structural basis for glycerol permeation by human AQP7
Sci Bull (Beijing), 66, 2020
|
|
6LNI
 
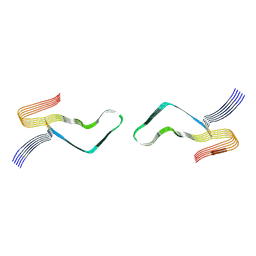 | | Cryo-EM structure of amyloid fibril formed by full-length human prion protein | | 分子名称: | Major prion protein | | 著者 | Wang, L.Q, Zhao, K, Yuan, H.Y, Wang, Q, Guan, Z.Y, Tao, J, Li, X.N, Hao, M.M, Chen, J, Zhang, D.L, Zhu, H.L, Yin, P, Liu, C, Liang, Y. | | 登録日 | 2019-12-30 | | 公開日 | 2020-06-10 | | 最終更新日 | 2020-06-24 | | 実験手法 | ELECTRON MICROSCOPY (2.702 Å) | | 主引用文献 | Cryo-EM structure of an amyloid fibril formed by full-length human prion protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4NWK
 
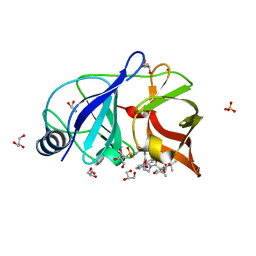 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-605339 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-n-((1r,2s)-1-((cyclopropylsulfonyl)carba moyl)-2-vinylcyclopropyl)-4-((6-methoxy-1-isoquinolinyl)ox y)-l-prolinamide | | 分子名称: | GLYCEROL, HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-4-[(6-methoxyisoquinolin-1-yl)oxy]-L-prolinamide, ... | | 著者 | Muckelbauer, J.K, Klei, H.E. | | 登録日 | 2013-12-06 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
4NWL
 
 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-650032 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-4-((7-chloro-4-methoxy-1-isoquinolinyl)o xy)-n-((1r,2s)-1-((cyclopropylsulfonyl)carbamoyl)-2-vinylc yclopropyl)-l-prolinamide | | 分子名称: | HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, ZINC ION | | 著者 | Muckelbauer, J.K, Klei, H.E. | | 登録日 | 2013-12-06 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
6JIF
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Pseudomonas sp. UW4 | | 分子名称: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Zheng, X, Guo, L, Li, D.F, Wu, B. | | 登録日 | 2019-02-21 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase from Pseudomonas sp. for efficient biosynthesis of chiral amino acids.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
6V6W
 
 | |
6KWY
 
 | | human PA200-20S complex | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, Proteasome activator complex subunit 4, Proteasome subunit alpha type-1, ... | | 著者 | Ouyang, S, Hongxin, G. | | 登録日 | 2019-09-09 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Plos Biol., 18, 2020
|
|
6KWX
 
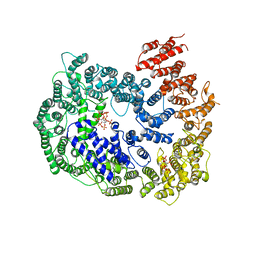 | | cryo-EM structure of human PA200 | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, Proteasome activator complex subunit 4, [(1~{S},2~{R},3~{R},4~{S},5~{S},6~{R})-2-[oxidanyl(phosphonooxy)phosphoryl]oxy-3,4,5,6-tetraphosphonooxy-cyclohexyl] phosphono hydrogen phosphate | | 著者 | Ouyang, S, Hongxin, G. | | 登録日 | 2019-09-09 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Plos Biol., 18, 2020
|
|
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | 著者 | Liu, Z, Yan, A, Gao, Y. | | 登録日 | 2022-10-20 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
8H7Z
 
 | |
1SVL
 
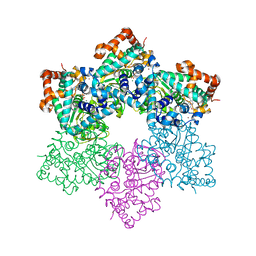 | | Co-crystal structure of SV40 large T antigen helicase domain and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | 著者 | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | 登録日 | 2004-03-29 | | 公開日 | 2004-10-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
1SVM
 
 | | Co-crystal structure of SV40 large T antigen helicase domain and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | 著者 | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | 登録日 | 2004-03-29 | | 公開日 | 2004-10-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
1SVO
 
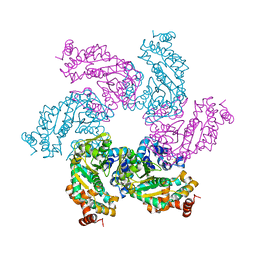 | | Structure of SV40 large T antigen helicase domain | | 分子名称: | ZINC ION, large T antigen | | 著者 | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | 登録日 | 2004-03-29 | | 公開日 | 2004-10-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
3VK6
 
 | |
5O6D
 
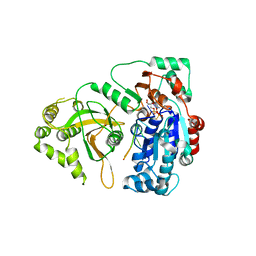 | | Structure of ScPif1 in complex with polydT and ATPgS | | 分子名称: | ATP-dependent DNA helicase PIF1, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | 著者 | Lu, K.Y, Chen, W.F, Rety, S, Liu, N.N, Xu, X.G. | | 登録日 | 2017-06-06 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.283 Å) | | 主引用文献 | Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Nucleic Acids Res., 46, 2018
|
|
