5UVC
 
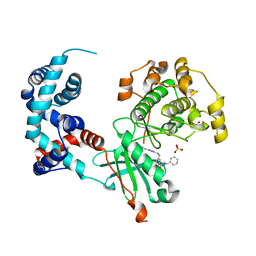 | | Design, Synthesis, and Evaluation of the First Selective and Potent G-protein-Coupled Receptor Kinase 2 (GRK2) Inhibitor for the Potential Treatment of Heart Failure | | Descriptor: | Beta-adrenergic receptor kinase 1, N-benzyl-3-({[5-(pyridin-4-yl)-4H-1,2,4-triazol-3-yl]methyl}amino)benzamide, SULFATE ION | | Authors: | Hoffman, I.D, Lawson, J.D. | | Deposit date: | 2017-02-20 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design, Synthesis, and Evaluation of the Highly Selective and Potent G-Protein-Coupled Receptor Kinase 2 (GRK2) Inhibitor for the Potential Treatment of Heart Failure.
J. Med. Chem., 60, 2017
|
|
5UUU
 
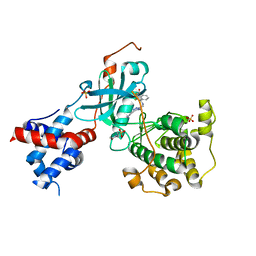 | | Design, Synthesis, and Evaluation of the First Selective and Potent G-protein-Coupled Receptor Kinase 2 (GRK2) Inhibitor for the Potential Treatment of Heart Failure | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-({[4-methyl-5-(pyridin-4-yl)-4H-1,2,4-triazol-3-yl]methyl}amino)-N-[2-(trifluoromethyl)benzyl]benzamide, Beta-adrenergic receptor kinase 1, ... | | Authors: | Hoffman, I.D, Lawson, J.D. | | Deposit date: | 2017-02-17 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, and Evaluation of the Highly Selective and Potent G-Protein-Coupled Receptor Kinase 2 (GRK2) Inhibitor for the Potential Treatment of Heart Failure.
J. Med. Chem., 60, 2017
|
|
3S6Y
 
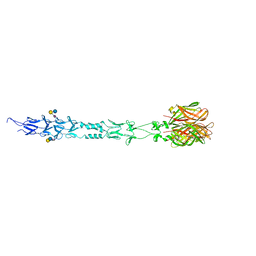 | | Structure of reovirus attachment protein sigma1 in complex with alpha-2,6-sialyllactose | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Outer capsid protein sigma-1 | | Authors: | Reiter, D.M, Dermody, T.S, Stehle, T. | | Deposit date: | 2011-05-26 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of reovirus attachment protein sigma1 in complex with sialylated oligosaccharides
Plos Pathog., 7, 2011
|
|
3S6Z
 
 | |
3S6X
 
 | |
4I3C
 
 | | Crystal structure of fluorescent protein UnaG N57Q mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bilirubin-inducible fluorescent protein UnaG | | Authors: | Kumagai, A, Ando, R, Miyatake, H, Miyawaki, A. | | Deposit date: | 2012-11-26 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | A bilirubin-inducible fluorescent protein from eel muscle
Cell(Cambridge,Mass.), 153, 2013
|
|
4I3B
 
 | | Crystal structure of fluorescent protein UnaG wild type | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bilirubin-inducible fluorescent protein UnaG, DI(HYDROXYETHYL)ETHER | | Authors: | Kumagai, A, Ando, R, Miyatake, H, Miyawaki, A. | | Deposit date: | 2012-11-26 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | A bilirubin-inducible fluorescent protein from eel muscle
Cell(Cambridge,Mass.), 153, 2013
|
|
4I3D
 
 | | Crystal structure of fluorescent protein UnaG N57A mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bilirubin-inducible fluorescent protein UnaG | | Authors: | Kumagai, A, Ando, R, Miyatake, H, Miyawaki, A. | | Deposit date: | 2012-11-26 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | A bilirubin-inducible fluorescent protein from eel muscle
Cell(Cambridge,Mass.), 153, 2013
|
|
6LK4
 
 | | Crystal structure of GMP reductase from Trypanosoma brucei in complex with guanosine 5'-triphosphate | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Guanosine 5'-monophosphate Reductase, PHOSPHATE ION | | Authors: | Mase, H, Otani, T, Imamura, A, Nishimura, S, Inui, T. | | Deposit date: | 2019-12-18 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Allosteric regulation accompanied by oligomeric state changes of Trypanosoma brucei GMP reductase through cystathionine-beta-synthase domain.
Nat Commun, 11, 2020
|
|
5XJM
 
 | | Complex structure of angiotensin II type 2 receptor with Fab | | Descriptor: | FabH, FabL, Sar1, ... | | Authors: | Asada, H, Horita, S, Shimamura, T, Iwata, S. | | Deposit date: | 2017-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog
Nat. Struct. Mol. Biol., 25, 2018
|
|
5XLI
 
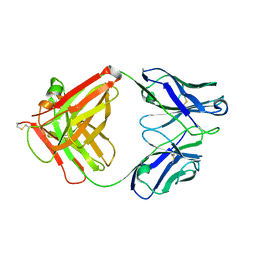 | | Structure of anti-Angiotensin II type2 receptor antibody (D5711-4A03) | | Descriptor: | FabH, FabL | | Authors: | Asada, H, Horita, S, Iwata, S, Hirata, K. | | Deposit date: | 2017-05-10 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6KS2
 
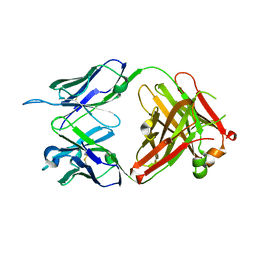 | | Structure of anti-Ghrelin receptor antibody | | Descriptor: | Fab7881 Heavy Chain (FabH), Fab7881 Light Chain (FabL) | | Authors: | Shiimura, Y, Horita, S, Asada, H, Hirata, K, Iwata, S, Kojima, M. | | Deposit date: | 2019-08-23 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structure of an antagonist-bound ghrelin receptor reveals possible ghrelin recognition mode.
Nat Commun, 11, 2020
|
|
6KO5
 
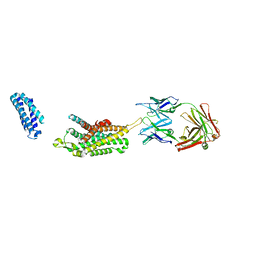 | | Complex structure of Ghrelin receptor with Fab | | Descriptor: | 6-(4-bromanyl-2-fluoranyl-phenoxy)-2-methyl-3-[[(3~{S})-1-propan-2-ylpiperidin-3-yl]methyl]pyrido[3,2-d]pyrimidin-4-one, Chimera of Soluble cytochrome b562 and Growth hormone secretagogue receptor type 1, Fab7881 Heavy Chain, ... | | Authors: | Shiimura, Y, Horita, S, Asada, H, Hirata, K, Iwata, S, Kojima, M. | | Deposit date: | 2019-08-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of an antagonist-bound ghrelin receptor reveals possible ghrelin recognition mode.
Nat Commun, 11, 2020
|
|
7W6Z
 
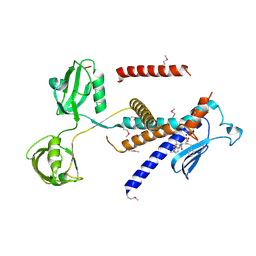 | |
7W6X
 
 | |
7W71
 
 | |
7W6Y
 
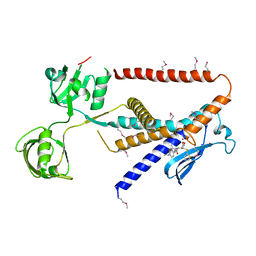 | | Crystal structure of Kangiella koreensis RseP orthologue in complex with batimastat in space group P1 | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, Anti sigma-E protein, RseA, ... | | Authors: | Imaizumi, Y, Takanuki, K, Nogi, T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanistic insights into intramembrane proteolysis by E. coli site-2 protease homolog RseP.
Sci Adv, 8, 2022
|
|
7W70
 
 | |
6NZV
 
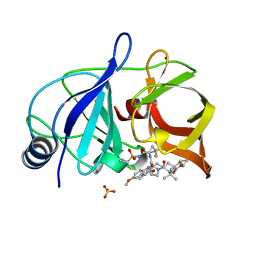 | | Crystal structure of HCV NS3/4A protease in complex with compound 12 | | Descriptor: | (1aR,5S,8S,9S,10R,22aR)-5-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-9-ethyl-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, HCV NS3/4A protease, SULFATE ION, ... | | Authors: | Appleby, T.C, Taylor, J.G. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of the pan-genotypic hepatitis C virus NS3/4A protease inhibitor voxilaprevir (GS-9857): A component of Vosevi®.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6NZT
 
 | |
7DB6
 
 | | human melatonin receptor MT1 - Gi1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Okamoto, H.H, Kusakizako, T, Shihioya, W, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-10-19 | | Release date: | 2021-08-18 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human MT 1 -G i signaling complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5EC5
 
 | | Crystal structure of lysenin pore | | Descriptor: | Lysenin, MERCURIBENZOIC ACID, MERCURY (II) ION | | Authors: | Podobnik, M, Savory, P, Rojko, N, Kisovec, M, Bruce, M, Jayasinghe, L, Anderluh, G. | | Deposit date: | 2015-10-20 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of an invertebrate cytolysin pore reveals unique properties and mechanism of assembly.
Nat Commun, 7, 2016
|
|
3UG2
 
 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with gefitinib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, Gefitinib | | Authors: | Parker, L.J, Handa, N, Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 32, 2013
|
|
3UG1
 
 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in the apo form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor | | Authors: | Parker, L.J, Handa, N, Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 32, 2013
|
|
3VJN
 
 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
