6J10
 
 | | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly | | Descriptor: | 6-cyclohexyl-4-methyl-1-oxidanyl-pyridin-2-one, Capsid protein | | Authors: | Park, S, Jin, M.S, Cho, Y, Kang, J, Kim, S, Park, M, Park, H, Kim, J, Park, S, Hwang, J, Kim, Y, Kim, Y.J. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly.
Nat Commun, 10, 2019
|
|
4MFU
 
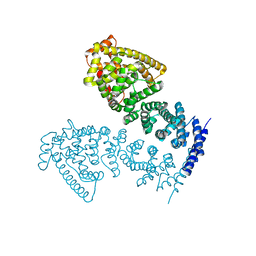 | | Crystal structure of human CTNNBL1(residues 77~563) | | Descriptor: | Beta-catenin-like protein 1 | | Authors: | Ahn, J.W, Kim, S, Kim, K.J. | | Deposit date: | 2013-08-28 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Structural insights into the novel ARM-repeat protein CTNNBL1 and its association with the hPrp19-CDC5L complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MFV
 
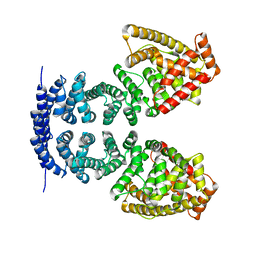 | | Crystal structure of human CTNNBL1(residues 33~563) | | Descriptor: | Beta-catenin-like protein 1 | | Authors: | Ahn, J.W, Kim, S, Kim, K.J. | | Deposit date: | 2013-08-28 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural insights into the novel ARM-repeat protein CTNNBL1 and its association with the hPrp19-CDC5L complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7JTB
 
 | |
7JSM
 
 | |
7JXA
 
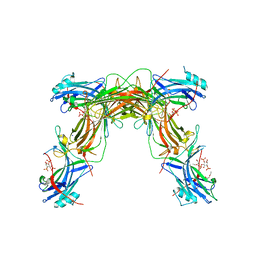 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH INOSITOL 1,4,5-TRIPHOSPHATE | | Descriptor: | 2-ETHOXYETHANOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, S-arrestin, ... | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
4J4L
 
 | |
7MP2
 
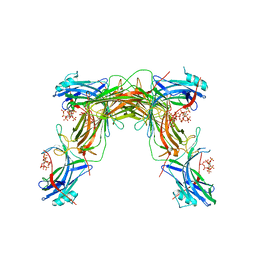 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1D-MYO-INOSITOL 1,5-BISDIPHOSPHATE TETRAKISPHOSPHATE (1,5-PP IP4) | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], S-arrestin | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7MOR
 
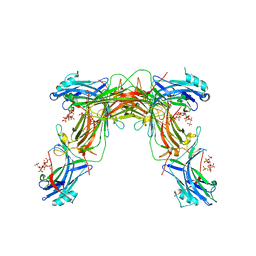 | |
7MP0
 
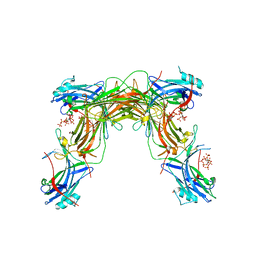 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1D-MYO-INOSITOL 5-DIPHOSPHATE PENTAKISPHOSPHATE (5-PP IP5) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, 2-ETHOXYETHANOL, S-arrestin | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7MP1
 
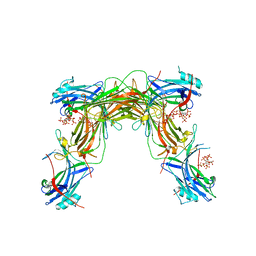 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1,5-DI-METHYLENEBISPHOSPHONATE INOSITOL TETRAKISPHOSPHATE (1,5-PCP-IP4) | | Descriptor: | S-arrestin, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(hydroxyphosphoryl)methanediyl]}bis(phosphonic acid) | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
5HZ2
 
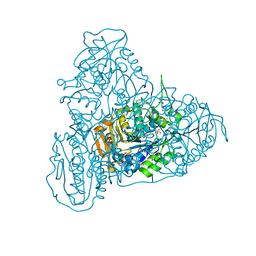 | | Crystal structure of PhaC1 from Ralstonia eutropha | | Descriptor: | GLYCEROL, Poly-beta-hydroxybutyrate polymerase, SULFATE ION | | Authors: | Kim, J, Kim, K.-J. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-07 | | Last modified: | 2017-04-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms.
Biotechnol J, 12, 2017
|
|
4IKC
 
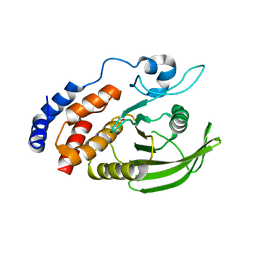 | | Crystal Structure of catalytic domain of PTPRQ | | Descriptor: | CHLORIDE ION, Phosphotidylinositol phosphatase PTPRQ, SULFATE ION | | Authors: | Yu, K.R, Ryu, S.E, Kim, S.J. | | Deposit date: | 2012-12-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis for the dephosphorylating activity of PTPRQ towards phosphatidylinositide substrates
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4TSV
 
 | |
7CYX
 
 | |
6KY2
 
 | | Crystal Structure of Arginine Kinase wild type from Daphnia magna | | Descriptor: | Arginine kinase, PHOSPHATE ION | | Authors: | Park, J.H, Rao, Z, Kim, S.Y, Kim, D.S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Insight into Structural Aspects of Histidine 284 of Daphnia magna Arginine Kinase.
Mol.Cells, 43, 2020
|
|
6KY3
 
 | | Structure of arginine kinase H284A mutant | | Descriptor: | ARGININE, Arginine kinase, PHOSPHATE ION, ... | | Authors: | Rao, Z, Park, J.H, Kim, S.Y, Kim, D.S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Insight into Structural Aspects of Histidine 284 of Daphnia magna Arginine Kinase.
Mol.Cells, 43, 2020
|
|
7CZ3
 
 | |
7BXA
 
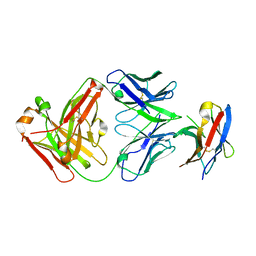 | | Crystal structure of PD-1 in complex with tislelizumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S, Lee, S.H, Lim, H, Lee, H.T, Kim, Y.J, Park, E.B. | | Deposit date: | 2020-04-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal structure of PD-1 in complex with an antibody-drug tislelizumab used in tumor immune checkpoint therapy.
Biochem.Biophys.Res.Commun., 527, 2020
|
|
5IN4
 
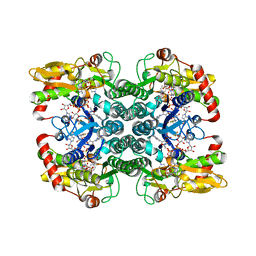 | | Crystal Structure of GDP-mannose 4,6 dehydratase bound to a GDP-fucose based inhibitor | | Descriptor: | GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Facile Modulation of Antibody Fucosylation with Small Molecule Fucostatin Inhibitors and Cocrystal Structure with GDP-Mannose 4,6-Dehydratase.
Acs Chem.Biol., 11, 2016
|
|
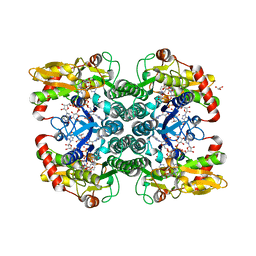
5IN5
 
 | |
2FLZ
 
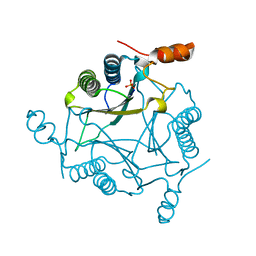 | |
2FLT
 
 | |
5Z7R
 
 | |
7LUV
 
 | | Cryo-EM structure of the yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2021-02-23 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast TREX complex and coordination with the SR-like protein Gbp2.
Elife, 10, 2021
|
|
