3LJ7
 
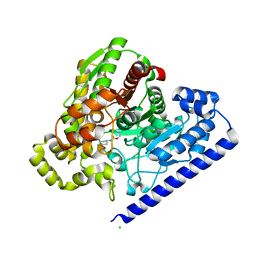 | |
1ZR4
 
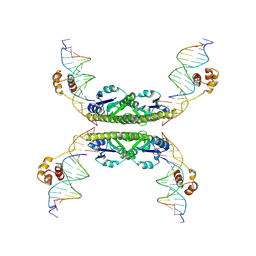 | | Structure of a Synaptic gamma-delta Resolvase Tetramer Covalently linked to two Cleaved DNAs | | Descriptor: | AAA, TCAGTGTCCGATAATTTAT, TTATCGGACACTG, ... | | Authors: | Li, W, Kamtekar, S, Xiong, Y, Sarkis, G.J, Grindley, N.D, Steitz, T.A. | | Deposit date: | 2005-05-19 | | Release date: | 2005-08-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs.
Science, 309, 2005
|
|
1ZR2
 
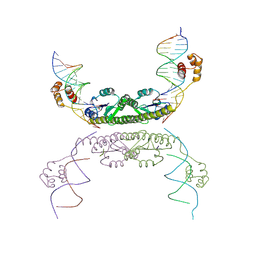 | | Structure of a Synaptic gamma-delta Resolvase Tetramer Covalently Linked to two Cleaved DNAs | | Descriptor: | AAA, TCAGTGTCCGATAATTTAT, TTATCGGACACTG, ... | | Authors: | Li, W, Kamtekar, S, Xiong, Y, Sarkis, G.J, Grindley, N.D, Steitz, T.A. | | Deposit date: | 2005-05-18 | | Release date: | 2005-08-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs.
Science, 309, 2005
|
|
2VYA
 
 | | Crystal Structure of fatty acid amide hydrolase conjugated with the drug-like inhibitor PF-750 | | Descriptor: | 4-(quinolin-3-ylmethyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Johnson, D.S, Wang, Z, Everdeen, D.S, Liimatta, M, Pabst, B, Bhattacharya, K, Nugent, R.A, Kamtekar, S, Cravatt, B.F, Ahn, K, Stevens, R.C. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Guided Inhibitor Design for Human Faah by Interspecies Active Site Conversion.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2PZS
 
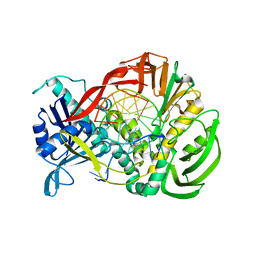 | | Phi29 DNA polymerase complexed with primer-template DNA (post-translocation binary complex) | | Descriptor: | 5'-d(CTAACACGTAAGCAGTC)-3', 5'-d(GACTGCTTAC)-3', DNA polymerase | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-18 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2PY5
 
 | | Phi29 DNA polymerase complexed with single-stranded DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-d(GGACTTT)-3', DNA polymerase | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-15 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2PYL
 
 | | Phi29 DNA polymerase complexed with primer-template DNA and incoming nucleotide substrates (ternary complex) | | Descriptor: | 1,2-ETHANEDIOL, 5'-d(CTGACGAATGTACA)-3', 5'-d(GACTGCTTAC(2DA))-3', ... | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2PYJ
 
 | | Phi29 DNA polymerase complexed with primer-template DNA and incoming nucleotide substrates (ternary complex) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-d(ACACGTAAGCAGTC)-3', ... | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
3LJ6
 
 | |
5UIS
 
 | | Crystal structure of IRAK4 in complex with compound 12 | | Descriptor: | 4-{[(3R)-piperidin-3-yl]oxy}-6-[(propan-2-yl)oxy]quinoline-7-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIQ
 
 | | Crystal structure of IRAK4 in complex with compound 9 | | Descriptor: | 2-[(propan-2-yl)oxy]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIR
 
 | | Crystal structure of IRAK4 in complex with compound 11 | | Descriptor: | 5-(4-cyanophenyl)-3-[(propan-2-yl)oxy]naphthalene-2-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIT
 
 | | Crystal structure of IRAK4 in complex with compound 14 | | Descriptor: | 1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}-7-[(propan-2-yl)oxy]isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIU
 
 | | Crystal structure of IRAK4 in complex with compound 30 | | Descriptor: | 1-{[(2S,3S,4S)-3-ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
2XRG
 
 | | Crystal structure of Autotaxin (ENPP2) in complex with the HA155 boronic acid inhibitor | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Hausmann, J, Albers, H.M.H.G, Perrakis, A. | | Deposit date: | 2010-09-14 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Substrate Discrimination and Integrin Binding by Autotaxin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3GNA
 
 | | Crystal structure of the RAG1 nonamer-binding domain with DNA | | Descriptor: | 5'-D(*AP*CP*TP*TP*AP*AP*CP*AP*AP*AP*AP*AP*CP*C)-3', 5'-D(*TP*GP*GP*TP*TP*TP*TP*TP*GP*TP*TP*AP*AP*G)-3', V(D)J recombination-activating protein 1 | | Authors: | Yin, F.F, Bailey, S, Innis, C.A, Steitz, T.A, Schatz, D.G. | | Deposit date: | 2009-03-16 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3GNB
 
 | | Crystal structure of the RAG1 nonamer-binding domain with DNA | | Descriptor: | 5'-D(*AP*AP*TP*TP*TP*TP*CP*AP*GP*AP*AP*AP*CP*C)-3', 5'-D(*AP*GP*GP*TP*TP*TP*CP*TP*GP*AP*AP*AP*AP*C)-3', V(D)J recombination-activating protein 1 | | Authors: | Yin, F.F, Bailey, S, Innis, C.A, Steitz, T.A, Schatz, D.G. | | Deposit date: | 2009-03-16 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis.
Nat.Struct.Mol.Biol., 16, 2009
|
|
5JI3
 
 | | HslUV complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
5JI2
 
 | | HslU L199Q in HslUV complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
1YYF
 
 | | Correction of X-ray Intensities from an HslV-HslU co-crystal containing lattice translocation defects | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent hsl protease ATP-binding subunit hslU, ATP-dependent protease hslV | | Authors: | Wang, J, Rho, S.H, Park, H.H, Eom, S.H. | | Deposit date: | 2005-02-24 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.16 Å) | | Cite: | Correction of X-ray intensities from an HslV-HslU co-crystal containing lattice-translocation defects.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
