6O87
 
 | |
6O88
 
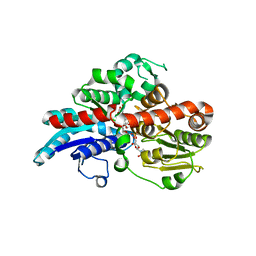 | | Crystal Structure of UDP-dependent glucosyltransferases (UGT) from Stevia rebaudiana in complex with UDP and rebaudioside A | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[alpha-L-allopyranosyl-(1->2)-[beta-D-mannopyranosyl-(1->3)]-beta-D-allopyranosyl]oxy}kauran-18-oic acid, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Lee, S.G, Jez, J.M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis for branched steviol glucoside biosynthesis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5MLM
 
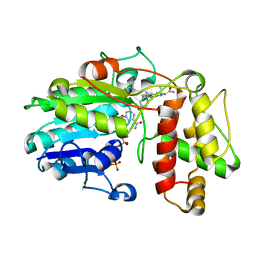 | | Plantago Major multifunctional oxidoreductase V150M mutant in complex with progesterone and NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, Progesterone 5-beta-reductase | | Authors: | Fellows, R, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2016-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.563 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
5MLH
 
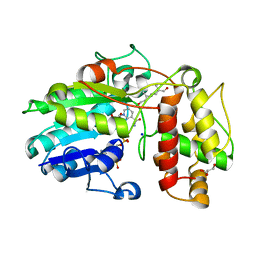 | | Plantago Major multifunctional oxidoreductase in complex with 8-oxogeranial and NADP+ | | Descriptor: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, CALCIUM ION, GLYCEROL, ... | | Authors: | Fellows, R, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2016-12-06 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
5MLR
 
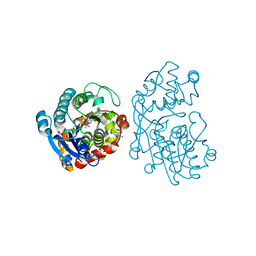 | | Plantago Major multifunctional oxidoreductase V150M mutant in complex with citral and NADP+ | | Descriptor: | CHLORIDE ION, Geranaldehyde, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Fellows, R, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2016-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
1QLV
 
 | |
7KQV
 
 | |
4EPM
 
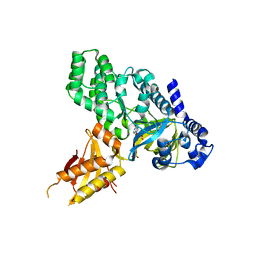 | | Crystal Structure of Arabidopsis GH3.12 (PBS3) in Complex with AMP | | Descriptor: | 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE, SULFATE ION | | Authors: | Westfall, C.S, Zubieta, C, Herrmann, J, Kapp, U, Nanao, M.H, Jez, J.M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
4EPL
 
 | | Crystal Structure of Arabidopsis thaliana GH3.11 (JAR1) in Complex with JA-Ile | | Descriptor: | Jasmonic acid-amido synthetase JAR1, N-({(1R,2R)-3-oxo-2-[(2Z)-pent-2-en-1-yl]cyclopentyl}acetyl)-L-isoleucine | | Authors: | Westfall, C.S, Zubieta, C, Herrmann, J, Kapp, U, Nanao, M.H, Jez, J.M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
6E1J
 
 | |
6E1Q
 
 | | AtGH3.15 acyl acid amido synthetase in complex with 2,4-DB | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, AtGH3.15 acyl acid amido synthetase, PHOSPHATE ION | | Authors: | Sharp, A.M, Lee, S.G, Jez, J.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Modification of auxinic phenoxyalkanoic acid herbicides by the acyl acid amido synthetase GH3.15 from Arabidopsis.
J. Biol. Chem., 293, 2018
|
|
6GSD
 
 | | Plantago Major multifunctional oxidoreductase in complex with progesterone and NADP+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROGESTERONE, Progesterone 5-beta-reductase | | Authors: | Fellows, R, Silva, C, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
5IUU
 
 | |
5IUW
 
 | | Crystal Structure of Indole-3-acetaldehyde Dehydrogenase in complexed with NAD+ and IAA | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Aldehyde dehydrogenase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, S.G, McClerklin, S, Kunkel, B, Jez, J.M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000.
PLoS Pathog., 14, 2018
|
|
5J34
 
 | | Isopropylmalate dehydrogenase K232M mutant | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, MAGNESIUM ION, ... | | Authors: | Lee, S.G, Jez, J.M. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | Structure and Mechanism of Isopropylmalate Dehydrogenase from Arabidopsis thaliana: INSIGHTS ON LEUCINE AND ALIPHATIC GLUCOSINOLATE BIOSYNTHESIS.
J.Biol.Chem., 291, 2016
|
|
5IUV
 
 | | Crystal Structure of Indole-3-acetaldehyde Dehydrogenase in complexed with NAD+ | | Descriptor: | Aldehyde dehydrogenase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, S.G, McClerklin, S, Kunkel, B, Jez, J.M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000.
PLoS Pathog., 14, 2018
|
|
3KAJ
 
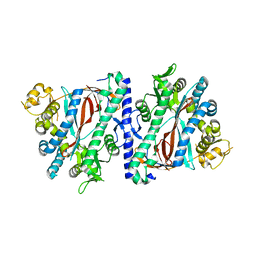 | | Apoenzyme structure of homoglutathione synthetase from Glycine max in open conformation | | Descriptor: | Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3KAK
 
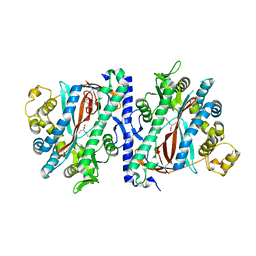 | | Structure of homoglutathione synthetase from Glycine max in open conformation with gamma-glutamyl-cysteine bound. | | Descriptor: | GAMMA-GLUTAMYLCYSTEINE, Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3KAL
 
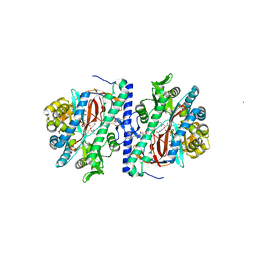 | | Structure of homoglutathione synthetase from Glycine max in closed conformation with homoglutathione, ADP, a sulfate ion, and three magnesium ions bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-gamma-glutamyl-L-cysteinyl-beta-alanine, MAGNESIUM ION, ... | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
4FGZ
 
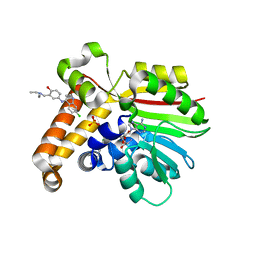 | | Crystal Structure of Phosphoethanolamine Methyltransferase from Plasmodium falciparum in Complex with Amodiaquine | | Descriptor: | 4-[(7-CHLOROQUINOLIN-4-YL)AMINO]-2-[(DIETHYLAMINO)METHYL]PHENOL, PHOSPHATE ION, Phosphoethanolamine N-methyltransferase, ... | | Authors: | Lee, S.G, Alpert, T.D, Jez, J.M. | | Deposit date: | 2012-06-05 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Crystal structure of phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with amodiaquine.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4FXP
 
 | |
4KRI
 
 | |
4KRG
 
 | |
1LWI
 
 | | 3-ALPHA-HYDROXYSTEROID/DIHYDRODIOL DEHYDROGENASE FROM RATTUS NORVEGICUS | | Descriptor: | 3-ALPHA-HYDROXYSTEROID/DIHYDRODIOL DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bennett, M.J, Schlegel, B.P, Jez, J.M, Penning, T.M, Lewis, M. | | Deposit date: | 1996-02-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase complexed with NADP+.
Biochemistry, 35, 1996
|
|
4KRH
 
 | |
