3WT1
 
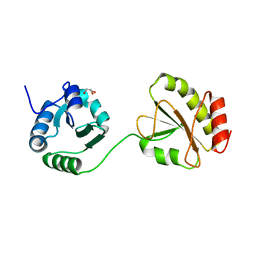 | | Crystal structure of the b'-a' domain of thermophilic fungal protein disulfide isomerase (reduced form) | | 分子名称: | GLYCEROL, Protein disulfide-isomerase | | 著者 | Inagaki, K, Satoh, T, Itoh, S.G, Okumura, H, Kato, K. | | 登録日 | 2014-04-02 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Redox-dependent conformational transition of catalytic domain of protein disulfide isomerase indicated by crystal structure-based molecular dynamics simulation
Chem.Phys.Lett., 618, 2015
|
|
3WT2
 
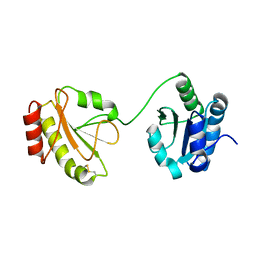 | | Crystal structure of the b'-a' domain of thermophilic fungal protein disulfide isomerase (oxidized form) | | 分子名称: | Protein disulfide-isomerase | | 著者 | Inagaki, K, Satoh, T, Itoh, S.G, Okumura, H, Kato, K. | | 登録日 | 2014-04-02 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Redox-dependent conformational transition of catalytic domain of protein disulfide isomerase indicated by crystal structure-based molecular dynamics simulation
Chem.Phys.Lett., 618, 2015
|
|
2RUE
 
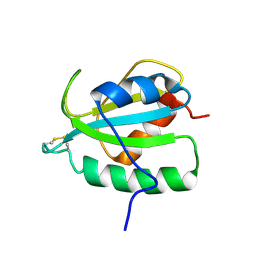 | |
2RUF
 
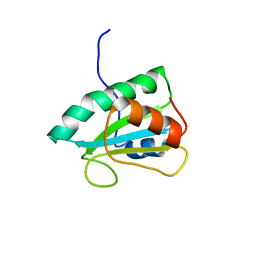 | |
1A05
 
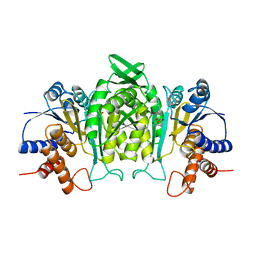 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THIOBACILLUS FERROOXIDANS WITH 3-ISOPROPYLMALATE | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MAGNESIUM ION | | 著者 | Imada, K, Inagaki, K, Matsunami, H, Kawaguchi, H, Tanaka, H, Tanaka, N, Namba, K. | | 登録日 | 1997-12-09 | | 公開日 | 1998-06-17 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure, 6, 1998
|
|
7F1U
 
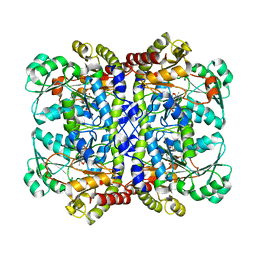 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-methionine intermediates | | 分子名称: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-(methylsulfanyl)but-2-enoic acid, L-methionine gamma-lyase, METHIONINE | | 著者 | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | 登録日 | 2021-06-09 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7F1P
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant ligand-free form. | | 分子名称: | L-methionine gamma-lyase | | 著者 | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | 登録日 | 2021-06-09 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7F1V
 
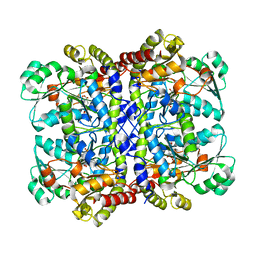 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-homocysteine intermediates | | 分子名称: | (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-sulfanyl-butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, L-methionine gamma-lyase | | 著者 | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | 登録日 | 2021-06-09 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
1GC2
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | 分子名称: | METHIONINE GAMMA-LYASE | | 著者 | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | 登録日 | 2000-07-06 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
1GC0
 
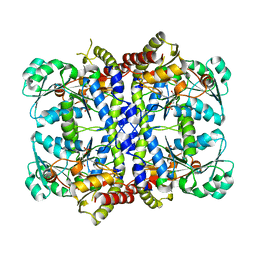 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | 分子名称: | METHIONINE GAMMA-LYASE | | 著者 | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | 登録日 | 2000-07-06 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
7CII
 
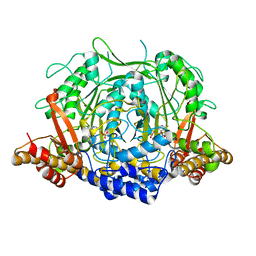 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with L- methionine methyl ester (external aldimine form). | | 分子名称: | L-methionine decarboxylase, methyl (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-4-methylsulfanyl-butanoate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIM
 
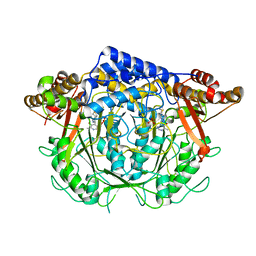 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with 3-methlythiopropylamine (geminal diamine form). | | 分子名称: | L-methionine decarboxylase, [6-methyl-4-[(3-methylsulfanylpropylamino)methyl]-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIJ
 
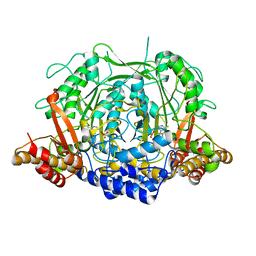 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with 3-methlythiopropylamine (external aldimine form). | | 分子名称: | L-methionine decarboxylase, [6-methyl-4-[(E)-3-methylsulfanylpropyliminomethyl]-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIG
 
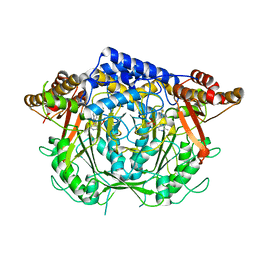 | | Crystal structure of L-methionine decarboxylase Q64A mutant from Streptomyces sp.590 in complexed with L- methionine methyl ester (geminal diamine form). | | 分子名称: | L-methionine decarboxylase, methyl (2S)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-methylsulfanyl-butanoate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIF
 
 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 (internal aldimine form). | | 分子名称: | L-methionine decarboxylase | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
3X0V
 
 | | Structure of L-lysine oxidase | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-lysine oxidase | | 著者 | Sano, T, Uchida, Y, Amano, M, Kawaguchi, T, Kondo, H, Inagaki, K, Imada, K. | | 登録日 | 2014-10-22 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Recombinant expression, molecular characterization and crystal structure of antitumor enzyme, l-lysine alpha-oxidase from Trichoderma viride.
J.Biochem., 157, 2015
|
|
7C3I
 
 | | Structure of L-lysine oxidase D212A/D315A | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | 登録日 | 2020-05-12 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
7C3L
 
 | | Structure of L-lysine oxidase D212A/D315A in complex with L-tyrosine | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-lysine oxidase, ... | | 著者 | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | 登録日 | 2020-05-12 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
7C3J
 
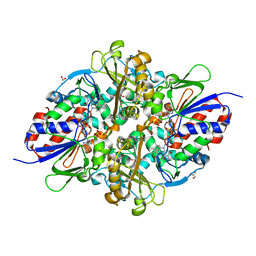 | | Structure of L-lysine oxidase D212A/D315A in complex with L-phenylalanine | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-lysine oxidase, ... | | 著者 | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | 登録日 | 2020-05-12 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
7D4C
 
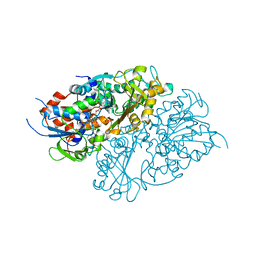 | | Structure of L-lysine oxidase precursor | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, PHOSPHATE ION | | 著者 | Ito, N, Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | 登録日 | 2020-09-23 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
7D4D
 
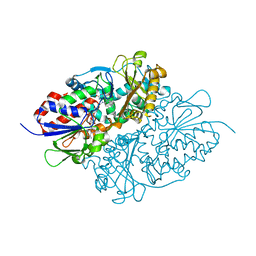 | | Structure of L-lysine oxidase precursor in complex with L-lysine (1.24M) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, LYSINE | | 著者 | Kitagawa, M, Ito, N, Matsumoto, Y, Inagaki, K, Imada, K. | | 登録日 | 2020-09-23 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
7D4E
 
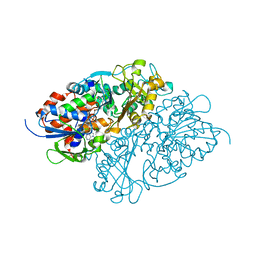 | | Structure of L-lysine oxidase precursor in complex with L-lysine (1.0 M) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, LYSINE | | 著者 | Kitagawa, M, Ito, N, Matsumoto, Y, Inagaki, K, Imada, K. | | 登録日 | 2020-09-23 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
7E0D
 
 | | Structure of L-glutamate oxidase R305E mutant in complex with L-arginine | | 分子名称: | ARGININE, FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase | | 著者 | Ito, N, Matsuo, S, Inagaki, K, Imada, K. | | 登録日 | 2021-01-27 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A new l-arginine oxidase engineered from l-glutamate oxidase.
Protein Sci., 30, 2021
|
|
7E0C
 
 | | Structure of L-glutamate oxidase R305E mutant | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase | | 著者 | Ito, N, Matsuo, S, Inagaki, K, Imada, K. | | 登録日 | 2021-01-27 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | A new l-arginine oxidase engineered from l-glutamate oxidase.
Protein Sci., 30, 2021
|
|
2D4V
 
 | | Crystal structure of NAD dependent isocitrate dehydrogenase from Acidithiobacillus thiooxidans | | 分子名称: | CITRATE ANION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, isocitrate dehydrogenase | | 著者 | Imada, K, Tamura, T, Namba, K, Inagaki, K. | | 登録日 | 2005-10-24 | | 公開日 | 2006-11-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: hydride transfer and co-factor specificity
Proteins, 70, 2008
|
|
