1L3B
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP W/ LONG CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1LUR
 
 | | Crystal Structure of the GalM/aldose Epimerase Homologue from C. elegans, Northeast Structural Genomics Target WR66 | | Descriptor: | SULFATE ION, aldose 1-epimerase | | Authors: | Keller, J.P, Xiao, R, MacDonald, L, Shen, J, Acton, T, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-05-23 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the GalM/aldose Epimerase Homologue from C. elegans, Northeast Structural Genomics Target WR66
TO BE PUBLISHED
|
|
1F38
 
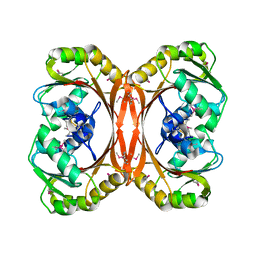 | |
1F35
 
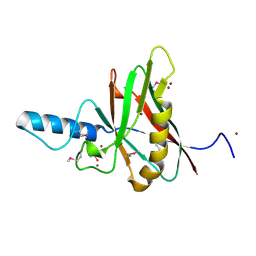 | |
1F3O
 
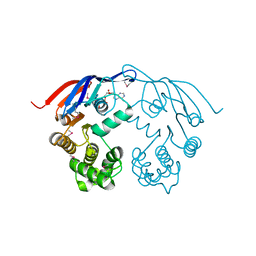 | | Crystal structure of MJ0796 ATP-binding cassette | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HYPOTHETICAL ABC TRANSPORTER ATP-BINDING PROTEIN MJ0796, MAGNESIUM ION | | Authors: | Yuan, Y.-R, Hunt, J.F. | | Deposit date: | 2000-06-05 | | Release date: | 2001-07-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the MJ0796 ATP-binding cassette. Implications for the structural consequences of ATP hydrolysis in the active site of an ABC transporter.
J.Biol.Chem., 276, 2001
|
|
1G9X
 
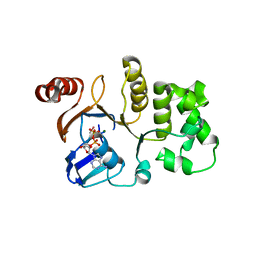 | | CHARACTERIZATION OF THE TWINNING STRUCTURE OF MJ1267, AN ATP-BINDING CASSETTE OF AN ABC TRANSPORTER | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HIGH-AFFINITY BRANCHED-CHAIN AMINO ACID TRANSPORT ATP-BINDING PROTEIN, MAGNESIUM ION, ... | | Authors: | Yuan, Y.-R, Hunt, J.F. | | Deposit date: | 2000-11-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of an MJ1267 ATP-binding cassette crystal with a complex pattern of twinning caused by promiscuous fiber packing.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1G6H
 
 | | CRYSTAL STRUCTURE OF THE ADP CONFORMATION OF MJ1267, AN ATP-BINDING CASSETTE OF AN ABC TRANSPORTER | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HIGH-AFFINITY BRANCHED-CHAIN AMINO ACID TRANSPORT ATP-BINDING PROTEIN, MAGNESIUM ION, ... | | Authors: | Yuan, Y.-R, Martsinkevich, O, Karpowich, N, Millen, L, Thomas, P.J, Hunt, J.F. | | Deposit date: | 2000-11-06 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter.
Structure, 9, 2001
|
|
1GAJ
 
 | | CRYSTAL STRUCTURE OF A NUCLEOTIDE-FREE ATP-BINDING CASSETTE FROM AN ABC TRANSPORTER | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HIGH-AFFINITY BRANCHED CHAIN AMINO ACID TRANSPORT ATP-BINDING PROTEIN, ... | | Authors: | Karpowich, N, Yuan, Y.-R, Dai, P.L, Martsinkevich, O, Millen, L, Thomas, P.J, Hunt, J.F. | | Deposit date: | 2000-11-30 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter.
Structure, 9, 2001
|
|
3RD6
 
 | | Crystal structure of Mll3558 protein from Rhizobium loti. Northeast Structural Genomics Consortium target id MlR403 | | Descriptor: | Mll3558 protein | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Ciccosanti, C, Sahdev, S, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-01 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Mll3558 protein from Rhizobium loti. Northeast Structural Genomics Consortium target id MlR403
To be Published
|
|
3RRU
 
 | | X-ray crystal structure of the VHS domain of human TOM1-like protein, Northeast Structural Genomics Consortium Target HR3050E | | Descriptor: | TOM1L1 protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, H, Ciccosanti, C, Patel, D, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-30 | | Release date: | 2011-06-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of the VHS domain of human TOM1-like protein, Northeast Structural Genomics Consortium Target HR3050E
To be Published
|
|
3RD4
 
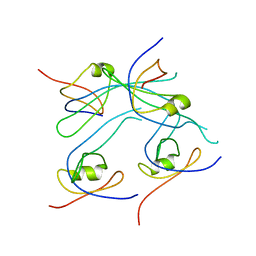 | | Crystal structure of PROPEN_03304 from Proteus penneri ATCC 35198 Northeast Structural Genomics Consortium target id PvR55 | | Descriptor: | uncharacterized protein | | Authors: | Seetharaman, J, Min, S, Kuzin, A, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of PROPEN_03304 from Proteus penneri ATCC 35198 Northeast Structural Genomics Consortium target id PvR55
To be Published
|
|
3SM3
 
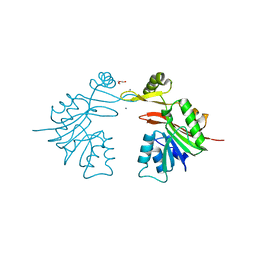 | | Crystal Structure of SAM-dependent methyltransferases Q8PUK2_METMA from Methanosarcina mazei. Northeast Structural Genomics Consortium Target MaR262. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, SAM-dependent methyltransferases | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Snyder, A, Patel, P, Xiao, R, Ciccosanti, C, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Crystal Structure of SAM-dependent methyltransferases Q8PUK2_METMA from Methanosarcina mazei.
To be Published, 2009
|
|
3SWV
 
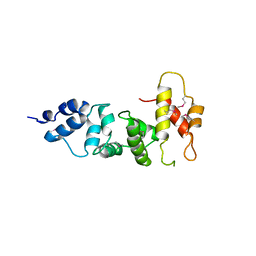 | | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A | | Descriptor: | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 | | Authors: | Seetharaman, J, Su, M, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-14 | | Release date: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BrefeldinA-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A
To be Published
|
|
3SY1
 
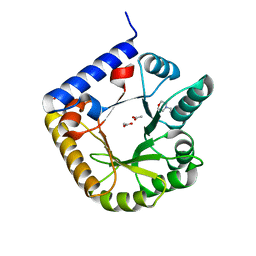 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR70 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, UPF0001 protein yggS | | Authors: | Vorobiev, S, Su, M, Nivon, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Maglaqui, M, Baker, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.465 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR70
To be Published
|
|
3SXW
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69. | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Vorobiev, S, Su, M, Nivon, L, Seetharaman, J, Patel, P, Xiao, R, Maglaqui, M, Ciccosanti, C, Baker, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69.
To be Published
|
|
3SYX
 
 | | Crystal Structure of the WH1 domain from human sprouty-related, EVH1 domain-containing protein. Northeast Structural Genomics Consortium Target HR5538B. | | Descriptor: | Sprouty-related, EVH1 domain-containing protein 1, YTTRIUM (III) ION | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Crystal Structure of the WH1 domain from human sprouty-related, EVH1 domain-containing protein.
To be Published
|
|
3TC6
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR63. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
3TC7
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | Descriptor: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
3THT
 
 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
3THP
 
 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
3U26
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR48 | | Descriptor: | PF00702 domain protein | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-23 | | Last modified: | 2022-03-02 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction.
Acs Chem.Biol., 8, 2013
|
|
3U02
 
 | | Crystal Structure of the tRNA modifier TiaS from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR225 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CITRIC ACID, ... | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-28 | | Release date: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal Structure of the tRNA modifier TiaS from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR225
To be Published
|
|
3U19
 
 | | CRYSTAL STRUCTURE OF ACYLENZYME INTERMEDIATE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | GLYCEROL, artificial protein OR51 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF ACYLENZYME INTERMEDIATE OF DE NOVO DESIGNED CYSTEINE
ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51
To be Published
|
|
3U13
 
 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3UAK
 
 | | Crystal Structure of De Novo designed cysteine esterase ECH14, Northeast Structural Genomics Consortium Target OR54 | | Descriptor: | De Novo designed cysteine esterase ECH14 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
